Twist Bioscience HQ
681 Gateway Blvd
South San Francisco, CA 94080
Methylation
- What modifications to the protocol can be made to optimize hybrid capture?
- What is my expected final library yield?
- Are there any internal controls to test for methylation conversion?
- What are the recommended number of amplification cycles post-capture?
- What is the min/max of DNA input for library preparation?
- Can I multiplex libraries in the capture; if so, what is the amount of library required?
- Should I use the Methylation Enhancer?
- What method of fragmentation can I use in this workflow?
- What is the recommended insert size?
- Can I use the Standard Hybridization workflow for enrichment?
- What is the final library size?
What modifications to the protocol can be made to optimize hybrid capture?
The following can be modified in order to optimize hybrid capture:
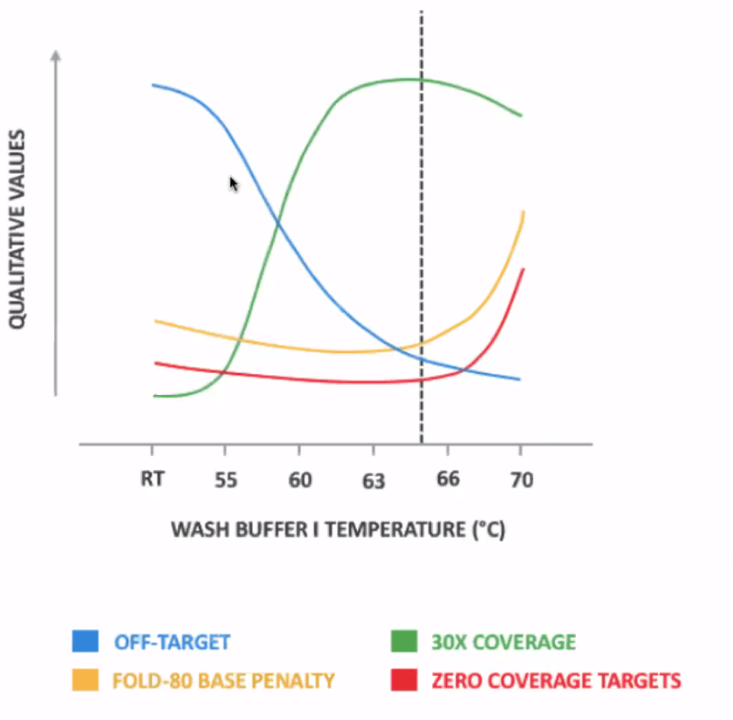
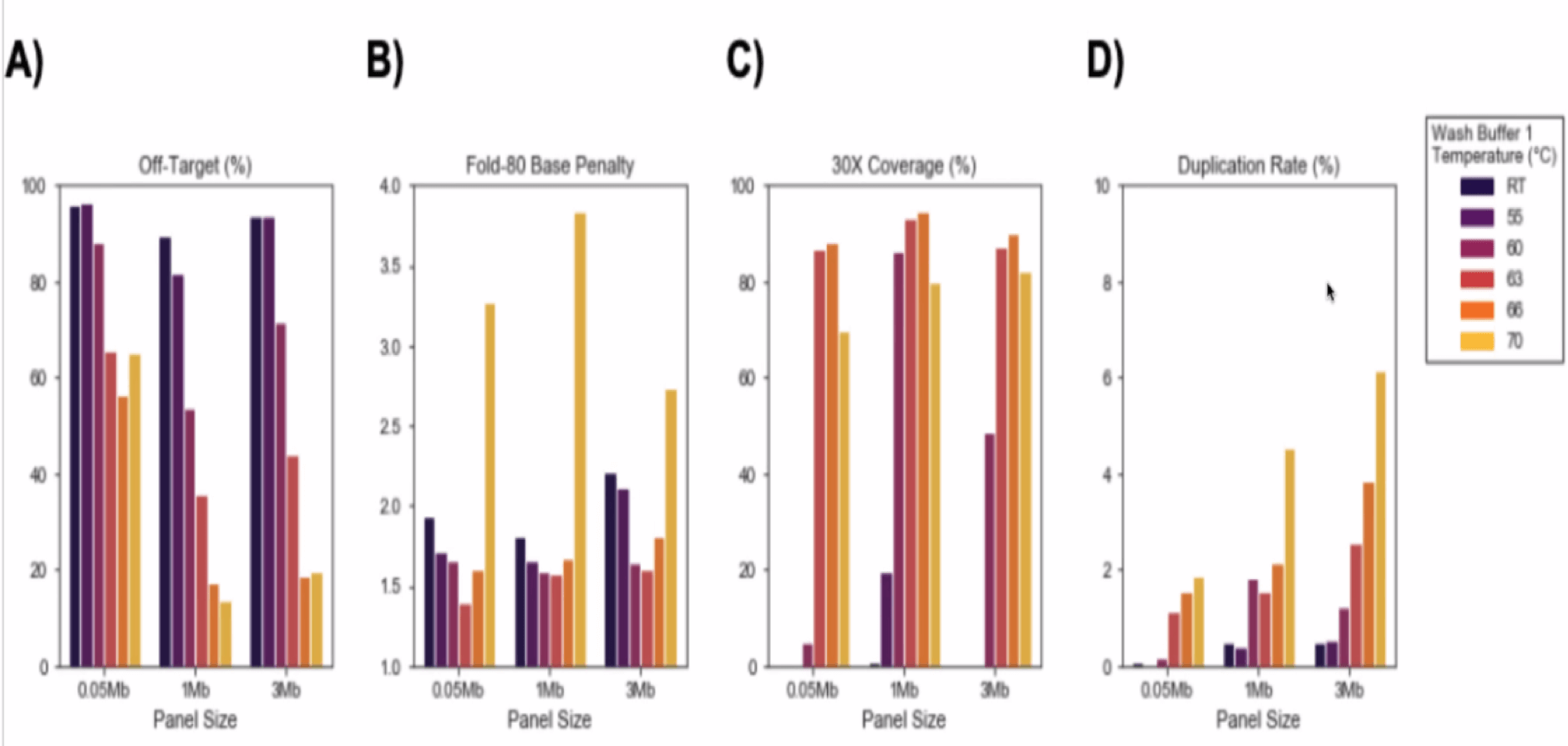
- Hybridization time – Start with 2 hours as recommended and modify between 30 min to 4 hours.
- Fast Wash Buffer 1 Temperature – Start with 65°C, adjust between 63-66°C:
Still have questions? Contact Us
What is my expected final library yield?
Expected final library yield is between 50-75ng/ul. The yield will depend on the quality of the starting input.
Still have questions? Contact Us
Are there any internal controls to test for methylation conversion?
CpG Methylated pUC19 DNA and Unmethylated Lambda DNA are included in the kit. These controls can be used to determine the efficiency of enzymatic conversion during library preparation.
NOTE: Internal controls should not be included in the capture workflow unless control-specific probes are added to the enrichment panel.
Still have questions? Contact Us
What are the recommended number of amplification cycles post-capture?
See table:
| Panel Size | Custom Panel | Custom Methylation Panel |
| >100 Mb | 5 | 8 |
| 50-100 Mb | 7 | 9 |
| 25-50 Mb | 8 | 10 |
| 10-25 Mb | 8 | 11 |
| 2.5-10 Mb | 9 | 12 |
| 1-2.5 Mb | 9 | 13 |
| 500-1,000 kb | 11 | 14 |
| 100-500 kb | 13 | 15 |
| 50-100 kb | 14 | 16 |
| <50 kb | 15 | 17 |
Still have questions? Contact Us
What is the min/max of DNA input for library preparation?
We recommend a minimum of 10-20 ng of high-quality DNA and maximum of 200 ng.
Still have questions? Contact Us
Can I multiplex libraries in the capture; if so, what is the amount of library required?
Yes, multiplexing up to an 8-plex is supported. We recommend using 200 ng of library for a single plex and 1500 ng (or 187.5 ng each) for an 8-plex capture.
Still have questions? Contact Us
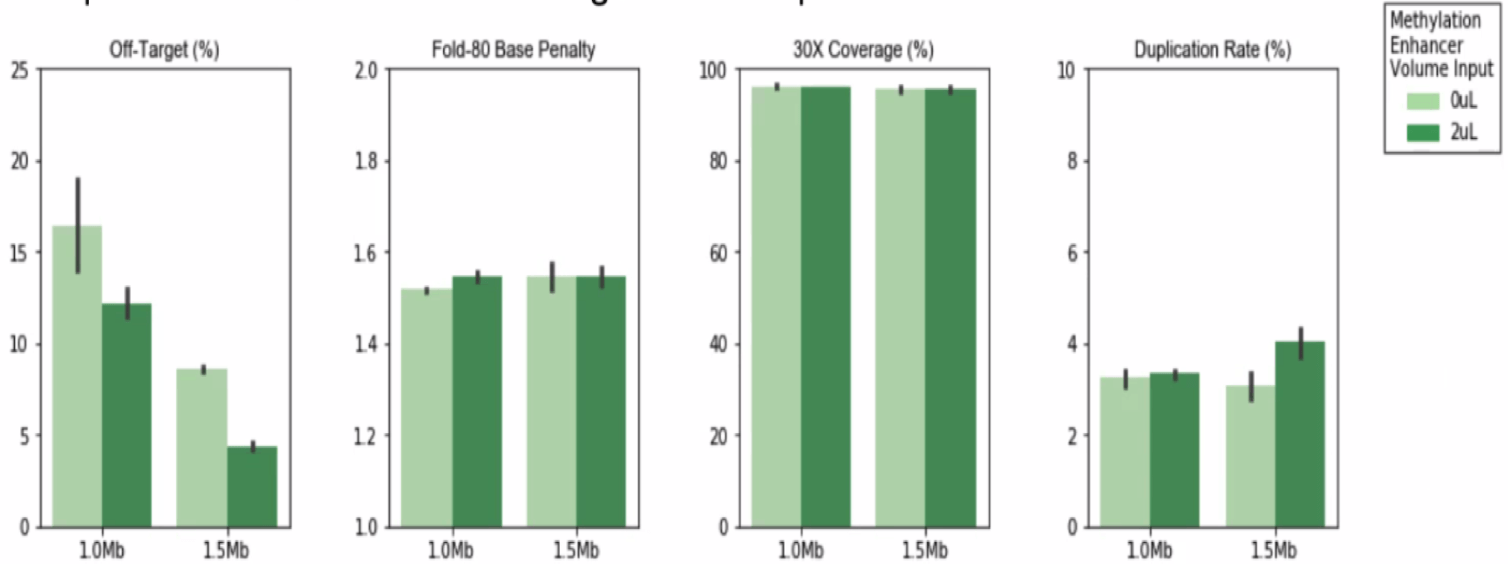
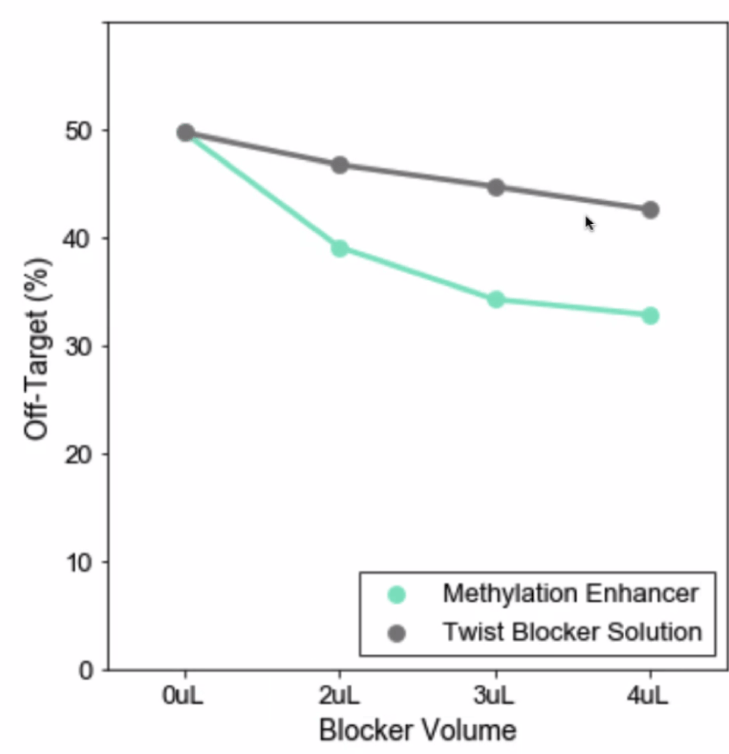
Should I use the Methylation Enhancer?
There is no detriment to any hybrid selection metrics when using this reagent. It reduces off-target in a variable fashion depending on custom methylation panel target regions and methylation states of the input genomic DNA. It can potentially decrease 50% in off-target in some custom panels.
Still have questions? Contact Us
What method of fragmentation can I use in this workflow?
Twist only supports Mechanical Fragmentation for this application when building libraries to undergo methylation conversion.
Still have questions? Contact Us
Can I use the Standard Hybridization workflow for enrichment?
No. Hybrid Capture must be performed with the Twist Fast Hybridization System.
Still have questions? Contact Us
What modifications to the protocol can be made to optimize hybrid capture?
The following can be modified in order to optimize hybrid capture:
- Hybridization time – Start with 2 hours as recommended and modify between 30 min to 4 hours.
- Fast Wash Buffer 1 Temperature – Start with 65°C, adjust between 63-66°C:


Still have questions? Contact Us
What is my expected final library yield?
Expected final library yield is between 50-75ng/ul. The yield will depend on the quality of the starting input.
Still have questions? Contact Us
Are there any internal controls to test for methylation conversion?
CpG Methylated pUC19 DNA and Unmethylated Lambda DNA are included in the kit. These controls can be used to determine the efficiency of enzymatic conversion during library preparation.
NOTE: Internal controls should not be included in the capture workflow unless control-specific probes are added to the enrichment panel.
Still have questions? Contact Us
What are the recommended number of amplification cycles post-capture?
See table:
| Panel Size | Custom Panel | Custom Methylation Panel |
| >100 Mb | 5 | 8 |
| 50-100 Mb | 7 | 9 |
| 25-50 Mb | 8 | 10 |
| 10-25 Mb | 8 | 11 |
| 2.5-10 Mb | 9 | 12 |
| 1-2.5 Mb | 9 | 13 |
| 500-1,000 kb | 11 | 14 |
| 100-500 kb | 13 | 15 |
| 50-100 kb | 14 | 16 |
| <50 kb | 15 | 17 |
Still have questions? Contact Us
What is the min/max of DNA input for library preparation?
We recommend a minimum of 10-20 ng of high-quality DNA and maximum of 200 ng.
Still have questions? Contact Us
Can I multiplex libraries in the capture; if so, what is the amount of library required?
Yes, multiplexing up to an 8-plex is supported. We recommend using 200 ng of library for a single plex and 1500 ng (or 187.5 ng each) for an 8-plex capture.
Still have questions? Contact Us
Should I use the Methylation Enhancer?
There is no detriment to any hybrid selection metrics when using this reagent. It reduces off-target in a variable fashion depending on custom methylation panel target regions and methylation states of the input genomic DNA. It can potentially decrease 50% in off-target in some custom panels.


Still have questions? Contact Us
What method of fragmentation can I use in this workflow?
Twist only supports Mechanical Fragmentation for this application when building libraries to undergo methylation conversion.
Still have questions? Contact Us
Can I use the Standard Hybridization workflow for enrichment?
No. Hybrid Capture must be performed with the Twist Fast Hybridization System.
Still have questions? Contact Us







