Precision Variant Library Technology Allows for Focused Screening
Twist’s massively parallel silicon-based DNA synthesis platform produces highly uniform and accurate oligos, with 90% of oligos represented within <2.5x of the mean, along with an industry-leading low error rate of 1:2,000 nt.
Combined with our well established molecular biology expertise, the Twist oligo synthesis platform enables the fabrication of highly diverse gene mutant libraries with excellent variant representation and highly specific user-defined composition with no unwanted bias or motifs. Twist library technology enables a comprehensive interrogation of the variant sequence space.
Learn more about Twist Bioscience’s ability to construct complex, high diversity libraries at confined regions (for example, CDRs) and introduce technology advancements that enable the construction of synthetic DNA libraries with diversity scattered along the length of constructs.
Learn how Twist helped a leading pharmaceutical company precisely interrogate key residues involved in both target recognition and antibody charge by synthesizing combinatorial variant libraries.
Precision Variant Library Technology Allows for Focused Screening
Twist’s massively parallel silicon-based DNA synthesis platform produces highly uniform and accurate oligos, with 90% of oligos represented within <2.5x of the mean, along with an industry-leading low error rate of 1:2,000 nt.
Combined with our well established molecular biology expertise, the Twist oligo synthesis platform enables the fabrication of highly diverse gene mutant libraries with excellent variant representation and highly specific user-defined composition with no unwanted bias or motifs. Twist library technology enables a comprehensive interrogation of the variant sequence space.
Learn more about Twist Bioscience’s ability to construct complex, high diversity libraries at confined regions (for example, CDRs) and introduce technology advancements that enable the construction of synthetic DNA libraries with diversity scattered along the length of constructs.
Learn how Twist helped a leading pharmaceutical company precisely interrogate key residues involved in both target recognition and antibody charge by synthesizing combinatorial variant libraries.
Twist Bioscience’s silicon-based DNA synthesis platform and library technology provides scientists with high-quality libraries that produce reliable data in less time. When compared to two other competing technologies (figure 1), Twist’s library showed less than 1% deviation from the designed amino acid frequency. In addition to precisely matching designed amino acid ratios, Twist’s in-silico DNA synthesis platform seamlessly incorporates desirable binding motifs and length variation across multi-domain libraries, giving scientists the power to precisely design and customize variant libraries that enable a comprehensive analysis of the variant space.
And with Twist libraries, the problems and challenges typical of NNK and TRIM libraries are avoided. Each variant is printed base-by-base and screened prior to synthesis, eliminating stop codons, liability motifs, unwanted mutations, and any undesirable biases, all at the beginning of the process. As a result, the library is enriched for the requested functional variants and the screening burden is reduced..
Our industry-leading, ready to use, highly-diverse and precisely designed libraries give scientists more opportunities to achieve their research goals.
At Twist, we use molecular biology expertise to precisely construct variant libraries. Our single-base control approach allows us to deliver high-diversity libraries without motifs that could confound your screening process. We deliver fully-customized libraries of unparalleled quality, with desired variants present at user-defined rations. Here you can see a CVL example representative of that quality. Variants in seven sequential amino acid positions were generated and all have expected variants at the positions shown, with nearly all at the desired ratio:
At position 1 and 6, the wild type amino acid was requested at 40% (position 1) and 30% (position 7). The remaining 18 amino acids were all requested to be low as 3.3%.
At position 3-5 amino acid residues were requested and observed at 5.3%.
Our Precise Variant Libraries allow you to choose what unique CDR (complimentary defining regions) sequences you want to be incorporated into the choice of framework(s).
Each CDR can be codon-optimized to avoid the creation of unwanted restriction sites. Machine learning has become an integral part of scientific research and has been used as a tool to analyze antibody libraries and identify unique CDR combinations that would yield, for example, higher affinity and specificity.
Coupled with Twist’s silicon-based synthesis platform, explicit library combinations generated from the analysis can be synthesized and seamlessly incorporated into a fully synthetic library to refine the exploration of the variant space.
Our platform enables uniform synthesis of highly complex oligonucleotides which minimizes potential bias in the downstream workflow. Our team of scientists have developed a strategy to maintain that uniformity throughout library fabrication and cloning. These are steps in the workflow that have been commonly shown to introduce bias which can lead to some variants showing up disproportionately in screens/assays. By maintaining the starting uniformity and minimizing the dropouts as well as under-represented variants, our cloned libraries are able to maintain a high level of diversity which helps reduce screening time and effort.
The figure shows the observed amino acid distribution at each variant position after fabrication (linear) and after cloning as it compares to the desired frequency (expected)
Since every Libraries verified with NGS, negative data can be used to identify mutations that do not yield improved functions, and those can be removed in the next iteration of library design.
The table shows the amino acid frequency (%) at four sites of a mutagenic domain after library linear assembly and after cloning. The expected columns for each position shows the desired frequency of each amino acid substitution. This data shows that all expected variants were present at all positions, and uniformity was maintained throughout assembly and cloning.
Twist Bioscience’s silicon-based DNA synthesis platform and library technology provides scientists with high-quality libraries that produce reliable data in less time. When compared to two other competing technologies (figure 1), Twist’s library showed less than 1% deviation from the designed amino acid frequency. In addition to precisely matching designed amino acid ratios, Twist’s in-silico DNA synthesis platform seamlessly incorporates desirable binding motifs and length variation across multi-domain libraries, giving scientists the power to precisely design and customize variant libraries that enable a comprehensive analysis of the variant space.
And with Twist libraries, the problems and challenges typical of NNK and TRIM libraries are avoided. Each variant is printed base-by-base and screened prior to synthesis, eliminating stop codons, liability motifs, unwanted mutations, and any undesirable biases, all at the beginning of the process. As a result, the library is enriched for the requested functional variants and the screening burden is reduced..
Our industry-leading, ready to use, highly-diverse and precisely designed libraries give scientists more opportunities to achieve their research goals.
At Twist, we use molecular biology expertise to precisely construct variant libraries. Our single-base control approach allows us to deliver high-diversity libraries without motifs that could confound your screening process. We deliver fully-customized libraries of unparalleled quality, with desired variants present at user-defined rations. Here you can see a CVL example representative of that quality. Variants in seven sequential amino acid positions were generated and all have expected variants at the positions shown, with nearly all at the desired ratio:
At position 1 and 6, the wild type amino acid was requested at 40% (position 1) and 30% (position 7). The remaining 18 amino acids were all requested to be low as 3.3%.
At position 3-5 amino acid residues were requested and observed at 5.3%.
Our Precise Variant Libraries allow you to choose what unique CDR (complimentary defining regions) sequences you want to be incorporated into the choice of framework(s).
Each CDR can be codon-optimized to avoid the creation of unwanted restriction sites. Machine learning has become an integral part of scientific research and has been used as a tool to analyze antibody libraries and identify unique CDR combinations that would yield, for example, higher affinity and specificity.
Coupled with Twist’s silicon-based synthesis platform, explicit library combinations generated from the analysis can be synthesized and seamlessly incorporated into a fully synthetic library to refine the exploration of the variant space.
Our platform enables uniform synthesis of highly complex oligonucleotides which minimizes potential bias in the downstream workflow. Our team of scientists have developed a strategy to maintain that uniformity throughout library fabrication and cloning. These are steps in the workflow that have been commonly shown to introduce bias which can lead to some variants showing up disproportionately in screens/assays. By maintaining the starting uniformity and minimizing the dropouts as well as under-represented variants, our cloned libraries are able to maintain a high level of diversity which helps reduce screening time and effort.
The figure shows the observed amino acid distribution at each variant position after fabrication (linear) and after cloning as it compares to the desired frequency (expected)
Since every Libraries verified with NGS, negative data can be used to identify mutations that do not yield improved functions, and those can be removed in the next iteration of library design.
The table shows the amino acid frequency (%) at four sites of a mutagenic domain after library linear assembly and after cloning. The expected columns for each position shows the desired frequency of each amino acid substitution. This data shows that all expected variants were present at all positions, and uniformity was maintained throughout assembly and cloning.
A New Way of Designing Your Libraries
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.



If you have any questions, please feel free to email us at [email protected]
Take a peek at our Library Design Tool
A New Way of Designing Your Libraries at Twist
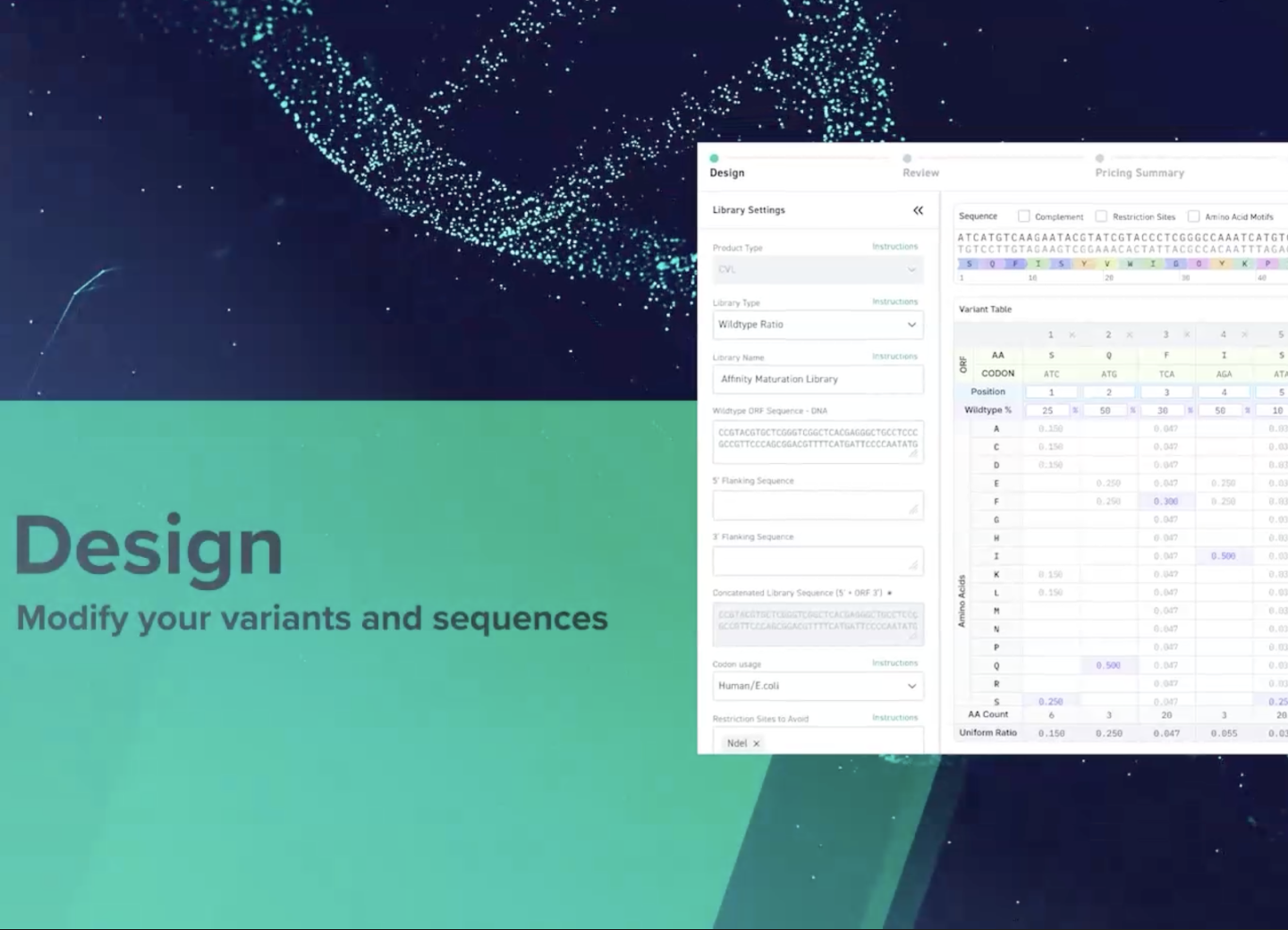
Watch Step by Step how to optimize your sequences
Let’s get Started
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
Note:
The Libraries Design Tool is not currently available in the APAC region. Please check with your local support team on availability date.
A New Way of Designing Your Libraries
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
If you have any questions, please feel free to email us at [email protected]



Take a peek at our Library Design Tool
A New Way of Designing Your Libraries at Twist

Watch Step by Step how to optimize your sequences
Let’s get Started
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
Note:
The Libraries Design Tool is not currently available in the APAC region. Please check with your local support team on availability date.