Generate 99% of Desired Variants
Protein engineering screens using single site variant libraries allow researchers to explore a protein’s sequence space and investigate the relationship between sequence and protein structure and function.
Twist Site Saturation Variant Library construction leverages massively parallel oligonucleotide synthesis using Twist's proprietary silicon-based DNA synthesis platform. Twist libraries are NGS-verified to confirm that all desired variants are present in the correct ratios.
Start Designing Your Library Now - A team of experts is waiting to help you design your perfect library for your specific needs. Order and Design here.
Learn more about how Twist is helping drug developers characterize and catalog mutations in KRAS through large-scale saturation mutagenesis screens.
Learn more about how Twist Single Site Variant Libraries (SSVL) enable the interrogation of sequence space to identify key residues in protein structure and function.
Learn how Twist is helping drug developers characterize and catalog mutations in KRAS through large-scale saturation mutagenesis screens.
Generate 99% of Desired Variants
Protein engineering screens using single site variant libraries allow researchers to explore a protein’s sequence space and investigate the relationship between sequence and protein structure and function.
Twist Site Saturation Variant Library construction leverages massively parallel oligonucleotide synthesis using Twist's proprietary silicon-based DNA synthesis platform. Twist libraries are NGS-verified to confirm that all desired variants are present in the correct ratios.
Learn more about how Twist is helping drug developers characterize and catalog mutations in KRAS through large-scale saturation mutagenesis screens.
Learn more about how Twist Single Site Variant Libraries (SSVL) enable the interrogation of sequence space to identify key residues in protein structure and function.
Start Designing Your Library Now - A team of experts is waiting to help you design your perfect library for your specific needs. Order and Design here.
Learn how Twist is helping drug developers characterize and catalog mutations in KRAS through large-scale saturation mutagenesis screens.
Site Saturation Libraries eliminate codon bias and unwanted substitutions unlike traditional methods such as degenerate and NNK approaches.
TRIM offers a poor repetitive yield, leading to <50% full-length product in a typical library whereas, Twist libraries yield more usable variants, increasing effective library size.
| Error Prone PCR | Degenerate
(NNK/NNS) |
Twist Site Saturation Variant Libraries |
|
|---|---|---|---|
| Eliminates sequence bias | No | No | Yes |
| Number of codons available | Unknown | 32 | All 64 |
| Prevents undesirable motifs | No | No | Yes |
| Allows codon optimization | No | No | Yes |
| Avoids stop codons | No | Yes | Yes |
Site Saturation Variant Libraries enable efficient sampling of a protein’s sequence space in screening assays.
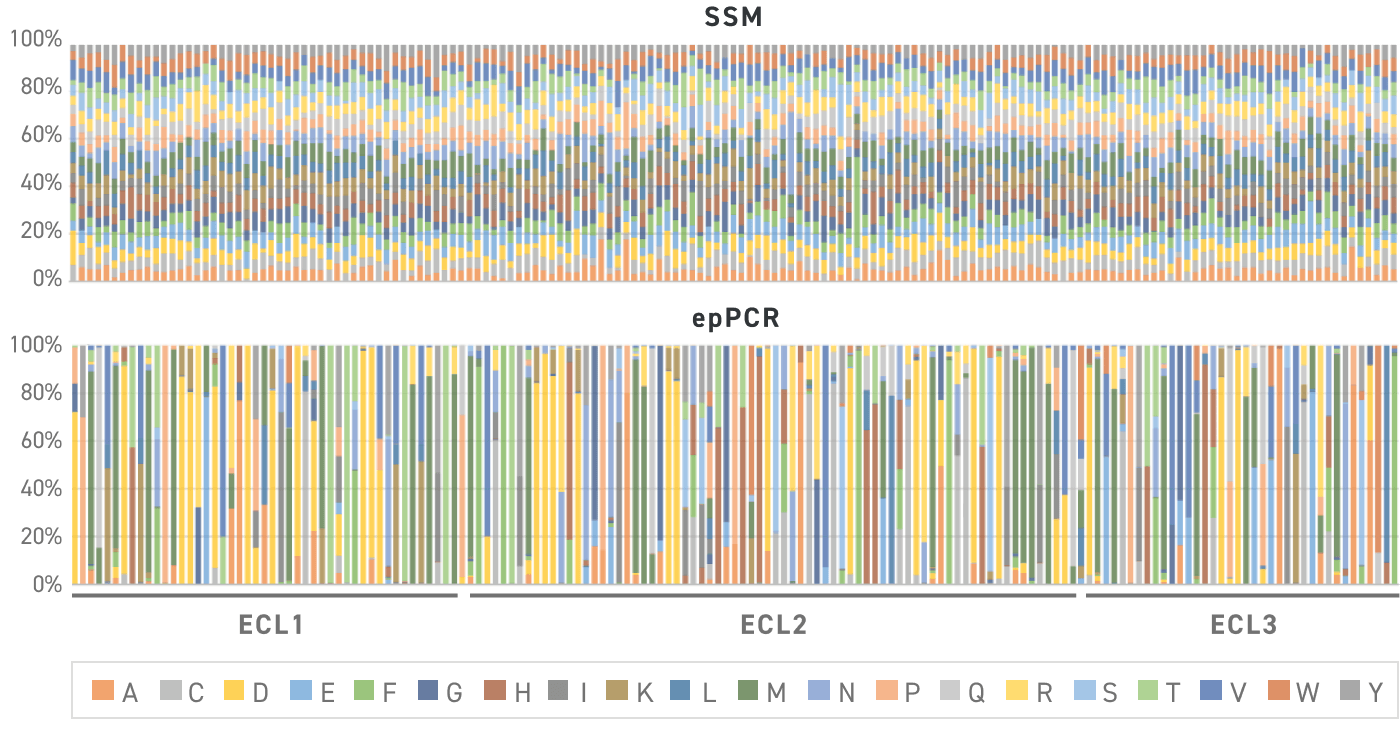
This figure shows the observed distribution of amino acids across 65 positions in protein (19 intended variants per position) when built with Twist SSVLs versus error prone PCR. The bars represent a different amino acid position, with each color indicating the observed variant frequency. All variants are present in the expected ratios in the Twist Library.
Site Saturation Libraries eliminate codon bias and unwanted substitutions unlike traditional methods such as degenerate and NNK approaches.
TRIM offers a poor repetitive yield, leading to <50% full-length product in a typical library whereas, Twist libraries yield more usable variants, increasing effective library size.
| Error Prone PCR | Degenerate
(NNK/NNS) |
Twist Site Saturation Variant Libraries |
|
|---|---|---|---|
| Eliminates sequence bias | No | No | Yes |
| Number of codons available | Unknown | 32 | All 64 |
| Prevents undesirable motifs | No | No | Yes |
| Allows codon optimization | No | No | Yes |
| Avoids stop codons | No | Yes | Yes |
Site Saturation Variant Libraries enable efficient sampling of a protein’s sequence space in screening assays.
This figure shows the observed distribution of amino acids across 65 positions in protein (19 intended variants per position) when built with Twist SSVLs versus error prone PCR. The bars represent a different amino acid position, with each color indicating the observed variant frequency. All variants are present in the expected ratios in the Twist Library.

A New Way of Designing Your Libraries
The New Library Design (DLD) Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.



If you have any questions, please feel free to email us at [email protected]
Take a peek at our Library Design Tool
A New Way of Designing Your Libraries at Twist
Watch Step by Step how to optimize your sequences
Let’s get Started
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
Note:
The Libraries Design Tool is not currently available in the APAC region. Please check with your local support team on availability date.
A New Way of Designing Your Libraries
The New Library Design (DLD) Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
If you have any questions, please feel free to email us at [email protected]



Take a peek at our Library Design Tool
A New Way of Designing Your Libraries at Twist

Watch Step by Step how to optimize your sequences
Let’s get Started
The New Library Design Tool is your all in one solution to streamline, optimize, and expedite the DNA Library design and ordering process.
Note:
The Libraries Design Tool is not currently available in the APAC region. Please check with your local support team on availability date.


