In the milieu of rushing blood, plasma, and cells, small wayward bits of DNA are easy to overlook. But stored within these scraps of genetic material, known as cell-free DNA (cfDNA), is information offering the earliest glimpses of cancer development. Being able to detect and understand these signals could dramatically improve our ability to detect and treat cancer before the disease has developed.
In contrast to healthy patients, where basal levels of cfDNA mostly derive from blood cells, patients with cancer show a measurable spike in cfDNA which can be traced back to the tumor microenvironment1. There, malignant and normal cells die at a high rate, dumping fragmented DNA from their genomes as they perish. CfDNA in the tumor microenvironment can also come from living cells that actively release bits of DNA in extracellular vesicles (we don’t yet know why living cells do this)1,2. Regardless, this release of tumor-derived cfDNA can happen very early in cancer development, giving researchers an opportunity to detect it and potentially prevent further progression.
There is a special term for cfDNA that derives from a tumor: circulating tumor DNA, or ctDNA. To differentiate ctDNA from the broader cfDNA circulating through a patient, scientists need to analyze the DNA fragments for specific features that are characteristic of tumors, such as the presence of cancer-causing mutations in the DNA sequence. One important feature that can tip off researchers that cancer is present is the pattern of methylation on the cfDNA.
Tracking cancer cells through DNA methylation
DNA methylation is what’s considered an epigenetic mark, meaning it’s genetic information that’s added on top of your DNA (epi is Greek for “above”). DNA methylation is one way in which cells control gene expression. DNA becomes methylated when enzymes, known as DNA methyltransferases, add a methyl group to cytosine bases, specifically to those cytosines that are positioned next to guanosines (this sequence is often referred to as CpG). Once attached, the methyl group acts as a buffer, blocking transcriptional proteins from interacting with the DNA3.
Methylated DNA therefore acts as a repressive coating that prevents genes from being expressed, allowing cells to temporarily turn off genes. This is similar to how you might archive data that you might not need for a long time. As such, genes that are heavily methylated are less likely to be expressed.
During development, cells begin to take on tissue-specific identities wherein they limit which genes are expressed, ensuring that liver cells, for example, behave like liver cells. Methylation is an important part of this process, which means that cells in each tissue all share a characteristic methylation pattern3.
When a cell becomes cancerous, it preserves some of that original methylation pattern. But, importantly, cancer cells also take on cancer-specific methylation patterns which allow them to turn on genes that help them persist and rapidly divide3-6. CfDNA therefore provides a unique window into cancer development. By looking for both tissue and cancer-specific patterns in cfDNA, researchers can determine if a person likely has cancer, and importantly in what tissue it’s developing.
🧬 Case Study: Detecting methylation patterns in cfDNA
Since 2012, Universal Diagnostics has been working to revolutionize early cancer detection. To make their high-throughput, NGS-based assay a reality, they needed a reputable, trustworthy source of custom panels for hybrid capture. They found the source they were looking for — as well as an enthusiastic research partner — in Twist Bioscience. Read more about this case study here >
cfDNA methyl-seq for cancer detection and characterization
While mutations in cfDNA can be detected using Next Generation Sequencing (NGS) technology, identifying methylation patterns on the DNA through methylation sequencing, or methyl-seq, is trickier.
Researchers have largely relied on a methyl-seq approach known as bisulfite sequencing wherein bisulfite salts chemically convert unmethylated cytosines into uracils. During amplification, uracils are converted to thymines. Subsequent parsing of the data can then determine which cytosines were methylated, and which weren’t.
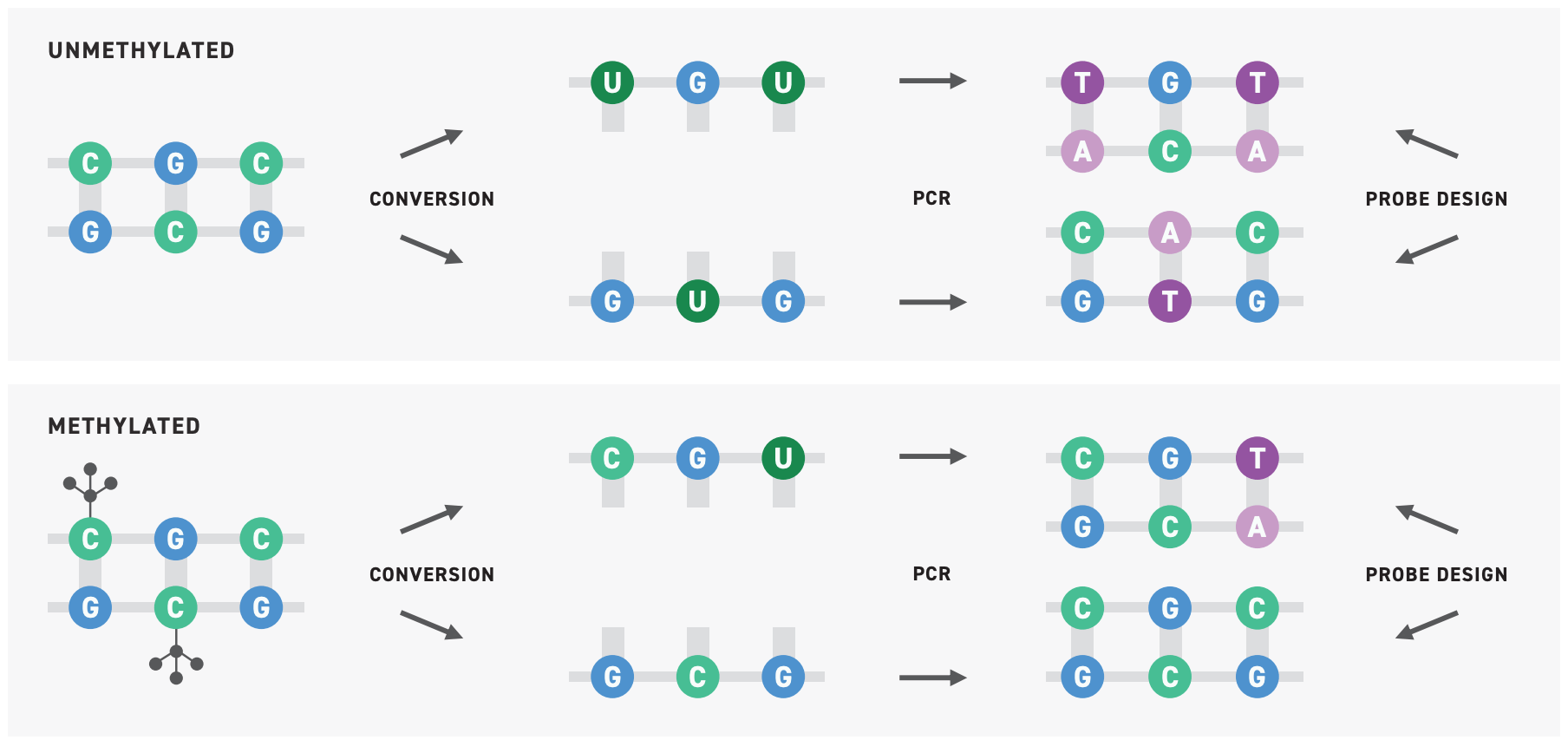
Bisulfite sequencing provides single base-pair resolution and has the potential for high-throughput applications, but it also substantially degrades DNA, decreasing the assay’s sensitivity, requiring large volumes of input DNA. This greatly limits its application in cfDNA methyl-seq.
To tap into methylation patterns within cfDNA, more advanced technology is needed.
Sensitive methyl-seq for cfDNA
Twist Bioscience has developed a new, highly sensitive method for detecting methylation patterns in cfDNA, known as the Twist NGS Methylation Detection System.
Leveraging New England Biolabs® innovative Enzymatic Methyl-seq (EM-seq™) technology, this system replaces bisulfite sequencing with a series of enzymatic conversions that ultimately convert unmethylated cytosine bases into uracil6. In relying on enzymes rather than bisulfite salts, the EM-seq protocol minimizes damage to the DNA and enables up to 15% more methylation detection compared to traditional bisulfite workflows.
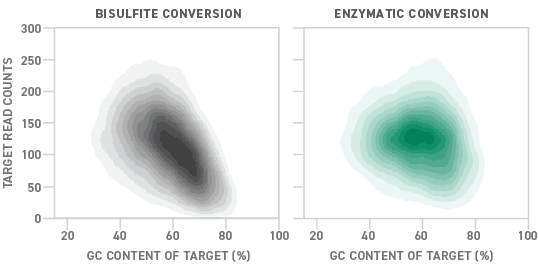
Following conversion, the sensitivity of the assay can be improved by target enrichment. However, as the conversion process involves flipping bases, it can both introduce novel DNA sequences that need to be anticipated, and reduce the DNA sequence’s complexity. If not accounted for, these can lead to high off-target rates and decreased sensitivity, both of which can be costly when working with cfDNA.
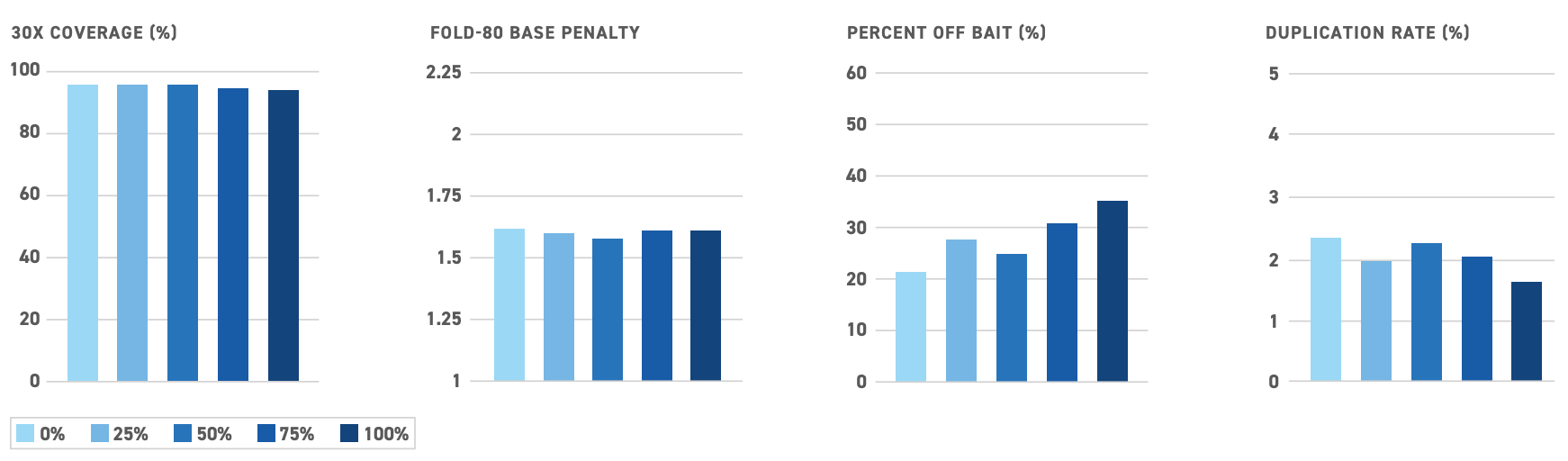
To improve assay performance, Twist has developed custom Methylation Panels that are optimized for methylation detection. These panels include probes to capture all four potential sequences at a given site: methylated, unmethylated, sense and antisense. And, as with all Twist Custom Panels, these Methylation Panels offer high fidelity, uniformity, and flexibility to allow for robust and sensitive methylation sequencing.
Even with expert support, designing a custom panel is far from straightforward. Sometimes, it makes more sense to go with an off-the-shelf solution, such as the Twist Human Methylome Panel. This highly developed target capture panel enriches 84% of CpG islands in the human genome, as well as several million additional CpG sites. When paired with Twist’s NGS Methylation Detection System, this panel can deliver highly uniform, precise, and broad methylation sequencing data.
Together, Twist’s methylation sequencing tools enable highly sensitive detection of methylation patterns in samples containing cfDNA.
EM-seq is a trademark of New England Biolabs, Inc.
Want to learn more about the Twist NGS Methylation Detection System?
Watch the short webinar below detailing the workflow, applications, and benefits of the Twist NGS Methylation Detection System.
References
- Bronkhorst, Abel Jacobus, et al. “The Emerging Role of Cell-Free DNA as a Molecular Marker for Cancer Management.” Biomolecular Detection and Quantification, vol. 17, Mar. 2019, p. 100087, 10.1016/j.bdq.2019.100087. Accessed 20 Sept. 2021.
- “The Main Sources of Circulating Cell-Free DNA: Apoptosis, Necrosis and Active Secretion.” Critical Reviews in Oncology/Hematology, vol. 157, 1 Jan. 2021, p. 103166, www.sciencedirect.com/science/article/pii/S1040842820303024, 10.1016/j.critrevonc.2020.103166. Accessed 20 Sept. 2021.
- Greenberg, Maxim V. C., and Deborah Bourc’his. “The Diverse Roles of DNA Methylation in Mammalian Development and Disease.” Nature Reviews Molecular Cell Biology, vol. 20, no. 10, 9 Aug. 2019, pp. 590–607, 10.1038/s41580-019-0159-6. Accessed 20 Sept. 2021.
- Zeng, Hu, et al. “Liquid Biopsies: DNA Methylation Analyses in Circulating Cell-Free DNA.” Journal of Genetics and Genomics, vol. 45, no. 4, Apr. 2018, pp. 185–192, 10.1016/j.jgg.2018.02.007. Accessed 23 Sept. 2021.
- Esteller, Manel. “Cancer Epigenomics: DNA Methylomes and Histone-Modification Maps.” Nature Reviews Genetics, vol. 8, no. 4, 6 Mar. 2007, pp. 286–298, 10.1038/nrg2005. Accessed 23 Sept. 2021.
- Vaisvila, Romualdas, et al. “Enzymatic Methyl Sequencing Detects DNA Methylation at Single-Base Resolution from Picograms of DNA.” Genome Research, 17 June 2021, genome.cshlp.org/content/early/2021/06/17/gr.266551.120, 10.1101/gr.266551.120. Accessed 30 Sept. 2021.