Improving crop yield. Eliminating Malaria in mosquitoes. Correcting previously incurable genetic diseases. Seven years after the debut of CRISPR-Cas9 as a genomic scissor, the question “what can Cas9 do?” has become difficult to answer. After all, what can’t Cas9 do? Remove its nuclease activity, and Cas9 becomes a modular platform for transcriptional control, epigenetic modification, genomic labeling, and base editing, among other applications. The tremendous biotechnological success of Cas9 has spurred efforts to expand the CRISPR toolbox to include other CRISPR nucleases, empowering the next generation of CRISPR applications.
CRISPR-associated (Cas) enzymes exist in most archaea and many bacteria as a component of their adaptive immunity. In 2015 — only a couple of years after Cas9 was first repurposed as a gene editor — Cas systems were organized into 2 classes, 5 types, and 16 subtypes based on their distinguishing features. Today, the classification system includes 2 classes, 6 types, and 33 subtypes (Makarova et al., 2020). This change was driven partly by the pursuit of new class 2 systems that could be repurposed for genome engineering à la Cas9. These include the DNA editor Cas12a (Cpf1) and Cas13-based RNA editors.
Argonautes, a group of nucleic acid-guided proteins mostly distinct from CRISPR-Cas systems, received bad press in 2016 when a high-profile paper was retracted after several independent groups failed to replicate claims of their so-called gene editing capabilities (Gao et al., 2016). Nevertheless, Argonautes warrant mention here, if only to point out an application for which they are better suited than CRISPR systems.
Below, we highlight a few of the most transformative developments in CRISPR (and Argonaute) technology compiled by Twist Bioscience’s experts.
Cas12 Simplifies Combinatorial Screens
Many biological processes such as tumor metastasis involve complex genetic interactions that cannot be fully resolved using a single-knockout approach. Although Cas9 is the gold-standard for single-knockout screening applications, its application in combinatorial screens has been hamstrung by its suboptimal multiplexing capabilities. Multiple independent groups recently optimized the type V CRISPR system Cas12a (Cpf1) to provide an improved alternative to Cas9 for combinatorial gene editing at scale (DeWeirdt et al., 2020; Gier et al., 2020).
What makes Cas12a better than Cas9 for combinatorial screens? Cas12a can process arrays of CRISPR RNAs (crRNAs) without RNase III and a trans-activating RNA (tracrRNA), components that are required for Cas9 to accomplish the same task. Because Cas9 cannot process crRNA arrays by itself, Cas9 guides typically must be expressed in separate expression cassettes, one per guide. The repetitive elements in multiplexed Cas9 guides make them prone to recombination and uncoupling during lentiviral delivery, PCR, and deep sequencing. These factors can complicate library design, cloning and analysis.
With Cas12a, oligos containing multiple crRNAs can be synthesized and easily cloned into lentiviral vectors for combinatorial screening applications. Moreover, the repetitive elements in multiplexed Cas12a guide vectors are comparatively short and more tolerant of sequence modifications, making multiplexed Cas12a guides less vulnerable to recombination and uncoupling throughout the screening process (DeWeirdt et al., 2020; Gier et al., 2020). A 300mer oligo from Twist Bioscience can encode 3-4 guides simultaneously.
While Cas12a is a great multiplexer, some of its limitations include a more restrictive protospacer adjacent motif (PAM) specificity (5’-TTTV) and lower mutation efficiency relative to Cas9. These parameters have been improved through structure-guided protein engineering of Cas12a from Acidaminococcus (AsCas12a; Kleinstiver et al., 2019), culminating in a modified enzyme named enhanced AsCas12a (enAsCas12a) that contains multiple point mutations in residues important for PAM recognition. In addition to expanding its PAM specificity (now including TTYN, VTTV, and TRTV, among others), enAsCas12a was serendipitously found to improve editing efficiency twofold on average compared to AsCas12a. AsCas12a’s efficiency has been further improved by adding multiple nuclear localization sequences and clarifying on- and off-target crRNA rules (DeWeirdt et al., 2020; Gier et al., 2020).
Type VI CRISPR Systems Set Their Sights on RNA
RNA interference (RNAi) using short-hairpin RNAs (shRNAs) is a well-established gene knockdown strategy that co-opts an endogenous RNA processing pathway to degrade cellular mRNAs. However, RNAi suffers from off-target effects and inconsistent knockdown efficiency. The specificity problem has been solved with CRISPR interference, a knockdown approach that uses a nuclease-dead Cas9 fused to the transcriptional repressor domain KRAB. Even so, both CRISPR interference and RNAi occasionally fail to robustly knock down their targets in cases of high basal gene expression or poor shRNA hairpin processing, respectively.
Enter RfxCas13d (CasRx), an RNA-guided, RNA-targeting CRISPR nuclease. CasRx may not have been the first RNA editor discovered, but side-by-side comparisons with other Cas13 systems (LwaCas13a and PspCas13b) and existing methods demonstrate that CasRx induces the most substantial knockdown across many genes — and with high specificity to boot. What’s more, a nuclease-dead variant dCasRx can be targeted to cis-elements of pre-mRNAs to regulate their splicing. To highlight this feature, Konermann et al. (2018) targeted dCasRx to an intronic splice silencer to rectify the pathological splicing of Tau-encoding mRNA in a neuronal model of frontotemporal dementia.
Because they prefer single-stranded RNA, the targeting of Cas13 systems in general can be hampered by RNA secondary structure (Abudayyeh et al., 2017). However, their ability to cause transient changes by targeting RNA over the Genome, and their lack of sequence constraints make them attractive transcriptome editing tools for both experimental and therapeutic applications.
Class 1 CRISPR-Cas Systems Diversify the CRISPR Toolkit
As class 2 CRISPR-Cas systems such as Cas9, Cas12s, and Cas13s continue to dominate the genome engineering space, class 1 CRISPR-Cas systems are beginning to garner attention because of their unique cleavage properties and multi-protein composition. The most common among these are type I systems, which combine the Cas3 helicase with a complex of accessory Cas proteins called CRISPR-associated complex for antiviral defense, or “Cascade”.
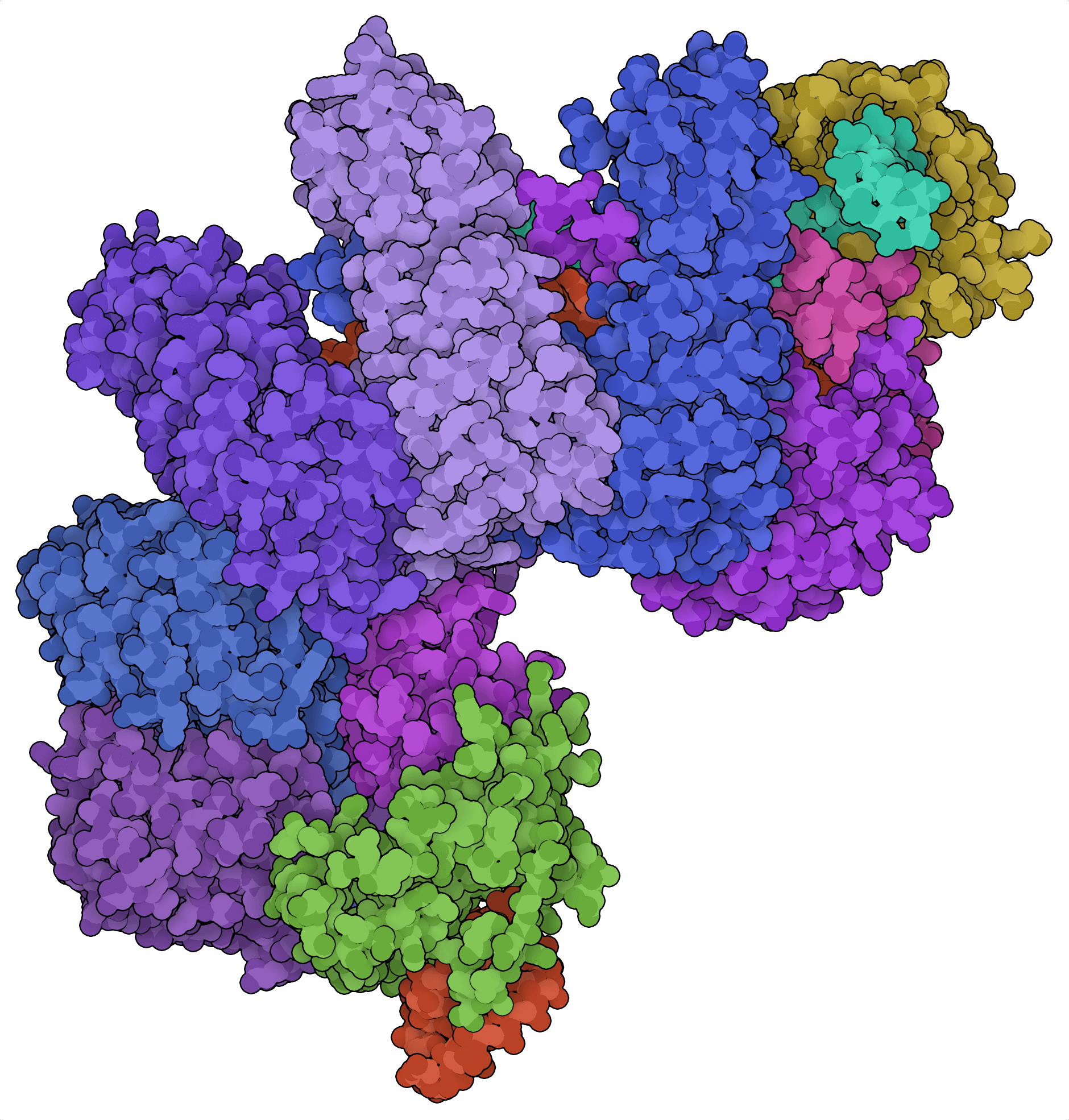
If Cas9 and Cas12a are genomic scissors, then Cas3 is a genomic shredder. Rather than producing small indels like Cas9, Cas3 causes a spectrum of large genomic deletions — ranging from 0.5 to 100 kb in length — upstream of targeted genomic sites (Dolan et al., 2019). This feature promises novel high-throughput screening applications whereby libraries of deletion mutants can be generated with comparatively fewer guides than class 2 CRISPR systems.
The Cascade complex alone also provides a modular platform for RNA-guided transcriptional activation and repression. Site-directed repression can be achieved using crRNAs in cas3-deleted E. coli (Luo et al., 2015; Rath et al., 2015) or in mammalian cells expressing exogenous Cascade proteins (Pickar-Oliver et al., 2019; Chen et al., 2020).
For gene activation, transcriptional activator domains need to be fused to one or more members of Cascade. Chen et al. (2020) obtained the best results when the synthetic VPR (VP64-p65-Rta) activator domain was fused to Csy3, the backbone component of Pseudomonas aeruginosa Cascade (PaeCascade) that interacts with the crRNA. Because multiple Csy3 proteins participate in the formation of the PaeCascade complex, multiple transcriptional effector domains can be recruited to a single Cascade complex. This strategy does not necessarily work for other class 1 systems, however, as the fusion of transcriptional effectors to Cas7 of EcoCascade (its Csy3 equivalent) failed to elevate transcription at targeted loci (Pickar-Oliver et al., 2019).
The multi-protein nature of Cascade comes in handy for building modular devices for transcriptional control, but it also means that multiple effector proteins must be co-expressed. This could impede the use of class 1 systems for some applications.
Strange CRISPR Loci Reveal CRISPR-associated Transposases
Most CRISPR loci contain genes for site-directed nucleases. For example, CRISPR loci encoding type I systems typically contain cas3, which encodes the endonuclease effector of these Cas systems. Recent bioinformatic analyses, however, revealed CRISPR loci containing Tn7-like transposon genes instead of Cas endonuclease genes. From these loci, two CRISPR-associated transposon systems have been characterized: the class 2 type V-K effector Cas12k (Strecker et al., 2019) and the class 1 type I-F Cascade complex (Halpin-Healy et al., 2020). Instead of cleaving DNA, these Cas systems can insert large segments of DNA into the genome in a site-directed and nuclease-independent fashion.
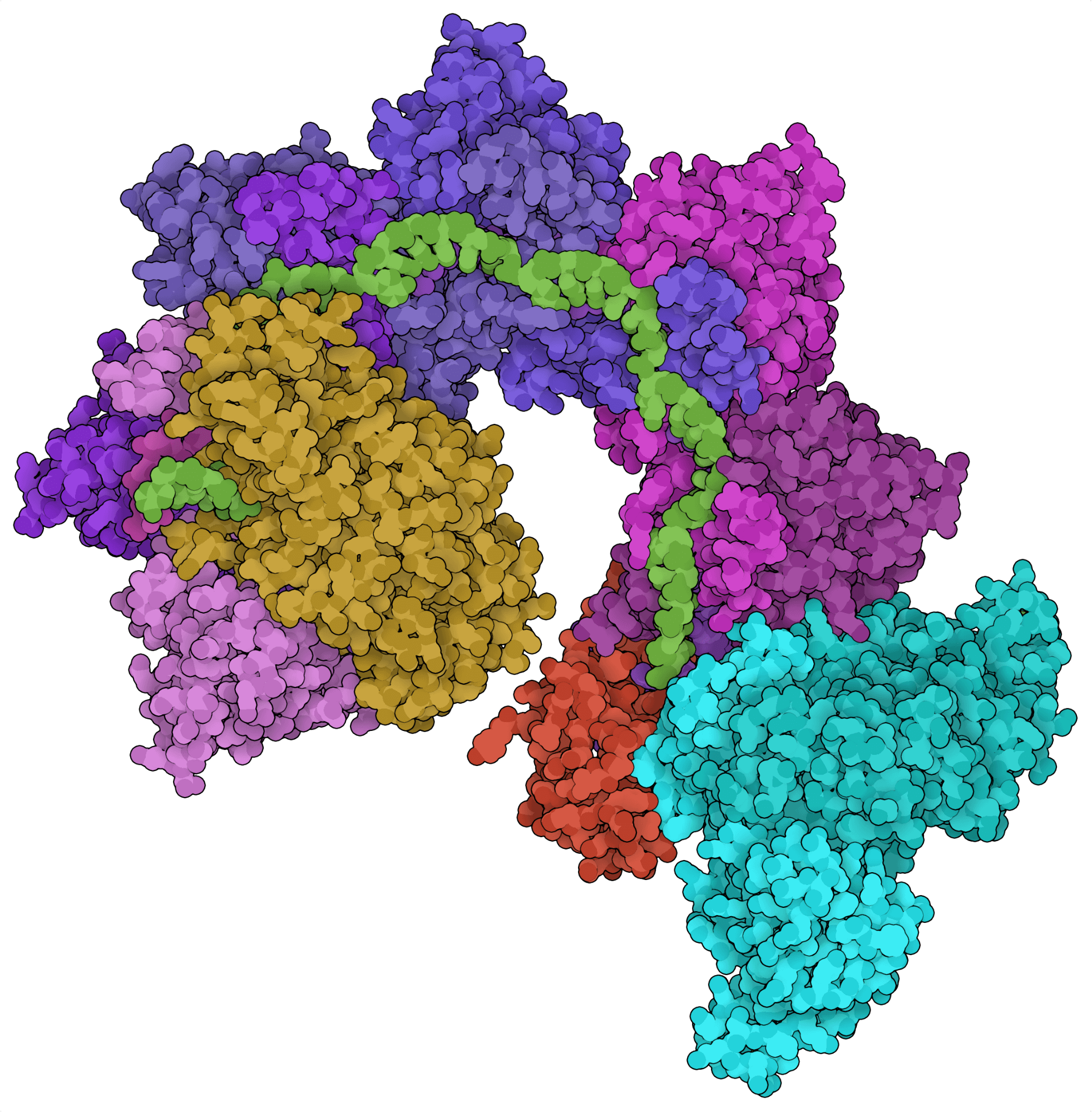
Traditional CRISPR-based gene knock-in approaches depend on homology-directed repair following a site-directed DNA break — a very inefficient process. The newly identified CRISPR-associated transposons offer a potentially more efficient workaround by allowing the transposition of large DNA fragments. CRISPR-associated transposases could be used to replace faulty exons or generate chimeric antigen receptor (CAR) T cells. While promising, the transposase activities of these systems have not yet been demonstrated in eukaryotic cells.
Argonautes Facilitate DNA Data Storage
Sometimes the best alternative to Cas9 is not a CRISPR system at all. This is true for nick-based DNA data storage, which requires a nicking enzyme to encode information in native DNA. DNA has been proposed as an alternative to current silicon-based storage media because of its relatively high durability and enormous storage density. To further push the limits of the information storage density of DNA, researchers have recently developed a nick-based approach (Tabatabaei et al., 2020).
Tabatabaei et al. (2020) leveraged the favorable nicking properties of PfArgo to create a DNA writing device that turns a dsDNA strand into a “punch card”. Here, binary information is encoded positionally in a pattern of nicks, which can be read by denaturing the DNA single-stranded fragments. These fragments can then be sequenced by next-generation or nanopore sequencing and mapped to the reference punch cards to retrieve the encoded information. Highlighting the outstanding storage density of this strategy, Tabatabaei et al. (2020) compressed a 329 kilobyte (KB) JPEG of the Lincoln Memorial into a 14 KB DNA file!
For nick-based DNA data storage, Cas9 nickase’s nicking properties pale in comparison to those of Pyrococcus furiosus Argonaute (PfAgo). Whereas the former is a single-turnover enzyme that relies on unstable and PAM-restricted RNA-DNA interactions for targeting, the latter can induce hundreds of nicks per enzyme using short DNA guides that are not limited by any specific targeting sequence.
These are just a few of the applications made possible by nucleic acid-guided proteins in the CRISPR and Argonaute families. To learn more about how to expand your CRISPR toolbox, talk with a member of the team.
Protein illustrations were generated from PDB structures using Illustrate, a non-photorealistic biomolecular illustration tool (Goodsell et al., 2019).
References
Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F (2017) RNA targeting with CRISPR-Cas13. Nature 550:280-284.
Chen Y, Liu J, Zhi S, Zheng Q, Ma W, Huang J, Liu Y, Liu D, Liang P, Songyang Z (2020) Repurposing type I-F CRISPR-Cas system as a transcriptional activation tool in human cells. Nat Commun 11:3136.
DeWeirdt, PC, Sangree, AK, Hanna, RE et al. (2020) Genetic screens in isogenic mammalian cell lines without single cell cloning. Nat Commun 11, 752.
Dolan AE, Hou Z, Xiao Y, Gramelspacher MJ, Heo J, Howden SE, Freddolino PL, Ke A, Zhang Y (2019) Introducing a Spectrum of Long-Range Genomic Deletions in Human Embryonic Stem Cells Using Type I CRISPR-Cas. Mol Cell 74:936-950 e935.
Gao F, Shen XZ, Jiang F, Wu Y, Han C (2016) DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nat Biotechnol 34:768-773.
Gier, RA, Budinich, KA, Evitt, NH et al. (2020) High-performance CRISPR-Cas12a genome editing for combinatorial genetic screening. Nat Commun 11, 3455
Goodsell, DS, Autin, L, Olson, AJ (2019) Illustrate: Software for Biomolecular Illustration. Structure 27:1716-1720.e1
Halpin-Healy TS, Klompe SE, Sternberg SH, Fernandez IS (2020) Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature 577:271-274.
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, Maus MV, Pinello L, Aryee MJ, Joung JK (2019) Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37:276-282.
Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018) Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 173:665-676 e614.
Luo ML, Mullis AS, Leenay RT, Beisel CL (2015) Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res 43:674-681.
Makarova KS et al. (2020) Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67-83.
Pickar-Oliver A, Black JB, Lewis MM, Mutchnick KJ, Klann TS, Gilcrest KA, Sitton MJ, Nelson CE, Barrera A, Bartelt LC, Reddy TE, Beisel CL, Barrangou R, Gersbach CA (2019) Targeted transcriptional modulation with type I CRISPR-Cas systems in human cells. Nat Biotechnol 37:1493-1501.
Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M (2015) Efficient programmable gene silencing by Cascade. Nucleic Acids Res 43:237-246.
Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F (2019) RNA-guided DNA insertion with CRISPR-associated transposases. Science 365:48-53.
Tabatabaei SK, Wang B, Athreya NBM, Enghiad B, Hernandez AG, Fields CJ, Leburton JP, Soloveichik D, Zhao H, Milenkovic O (2020) DNA punch cards for storing data on native DNA sequences via enzymatic nicking. Nat Commun 11:1742.