Researchers Unlock Secrets of Enzyme Blocking HIV Using Twist Bioscience’s Oligo Pools
Although the human immunodeficiency virus (HIV) can be controlled by antiviral therapy, there is no complete cure for the disease as of yet. HIV still affects more than 35 million people today. However, our bodies’ have developed a number of tools to protect us against viruses. Recent research has shown that HIV has evolved a number of tools that allow it to evade our bodies’ defense systems. The hope is that by better understanding the disease, we can be better armed to fight, cure and prevent it.
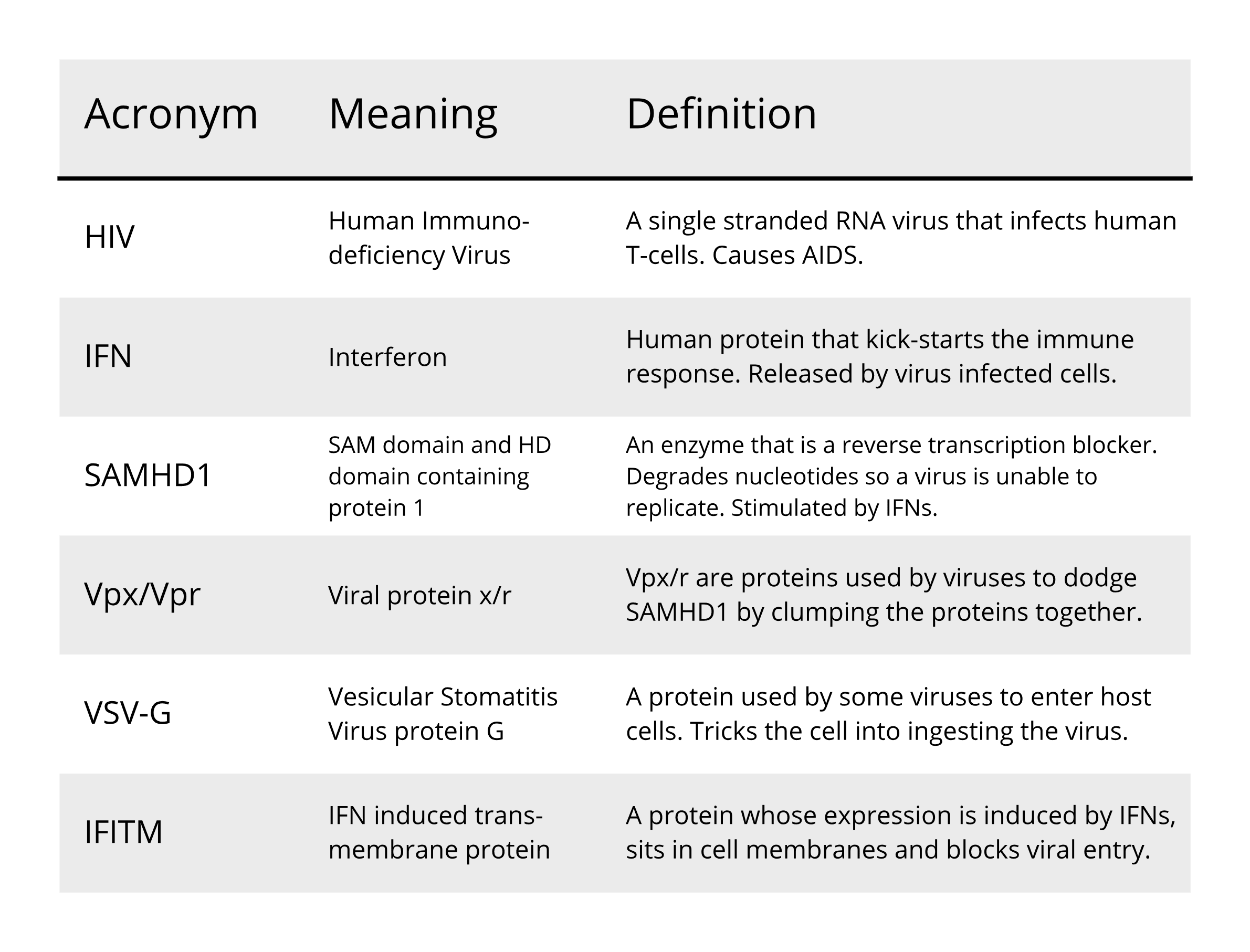
(Since virology employs a great deal of acronyms that we will also be using in this blog, we have included at the bottom of this piece a quick guide to the terms used for your reference).
HIV is a retrovirus. This means its genome is made up of a single strand of RNA. Once in the body, it infects and destroys T-cells in a bid to replicate itself.
However, our bodies are in a continuous arms race with these viruses. When our immune systems spot a viral intruder, an immune defense system kicks into action. On the front-line of this system is a set of proteins called type-1 interferons (IFNs). These kickstart the innate immune response, stimulating genes which block various aspects of the viral life cycle.
One of these proteins is the enzyme SAMHD1. When replicating, the virus needs nucleotide molecules to copy its genome. SAMHD1 counters this by rapidly depleting the available pool of nucleotides, halting replication. However, as HIV cannot persist unless it has a human host, it is under a huge evolutionary pressure to counter these mechanisms.
HIV viruses have evolved countermeasures to the SAMHD1 defense system, one of two proteins called Vpx and Vpr respectively. The virus spits one of these proteins into the infected cell alongside its own genetic material when it joins with the cell-membrane. Vpx and Vpr act as infiltrators, sending a message to the cell that tricks it into degrading its own SAMHD1.
Recent research has uncovered yet another layer to this battle. If cells are treated with a family of interferons called IFNα, SAMHD1 degradation is blocked, allowing nucleotide pool depletion to persist. In a recent report in Retrovirology, researchers gained valuable knowledge about how IFNα protects SAMHD1 from degradation, an experiment powered by CRISPR and oligonucleotide pools from Twist Bioscience.
The researchers took advantage of the high throughput nature of a technique called flow cytometry to assay the effects that interferon proteins have on SAMHD1 degradation. Instead of using real viruses, they infected cells with a lab-model of a virus called a virus-like particle. These particles contained Vpx and were coated in a protein called VSV-G. VSV-G is a virus-derived protein that allows the virus to combine with the cell membrane and dump its contents into the cell. HIV viruses have their own version of this protein and understanding differences between the two may be important for future research on HIV infection.
Using these virus-like particles, the researchers confirmed that in the presence of IFNα, lab-grown T-cells produce a number of proteins in their cell surface called IFN Induced Transmembrane (IFITM) proteins. These proteins sit in the membrane of the T-cell acting as guards. They restrict the entry of the virus-like particles into the cell, meaning SAMHD1 expression could persist as Vpx is also blocked.
The researchers then used a CRISPR-Cas9 knockout screen, combined with flow cytometry, to better understand which IFITM proteins were effective at blocking viral entry. The CRISPR screen was designed to target and knock out the expression 1906 genes, whose expression is known to be induced by IFNs, using a pool of 15448 gRNAs, encoded within an oligonucleotide pool that was synthesised by Twist Bioscience. The sgRNA library was synthesized in vitro and cloned in the lentiCRISPRv2 lentiviral backbone. Lab-grown T-cells were transduced with this library and cultured for two weeks to allow gene knockout to occur. The cells were then treated with IFNα, and incubated with virus-like particles. SAMHD1 expression levels were measured and cells were sorted into low and high expression pools using flow cytometry.
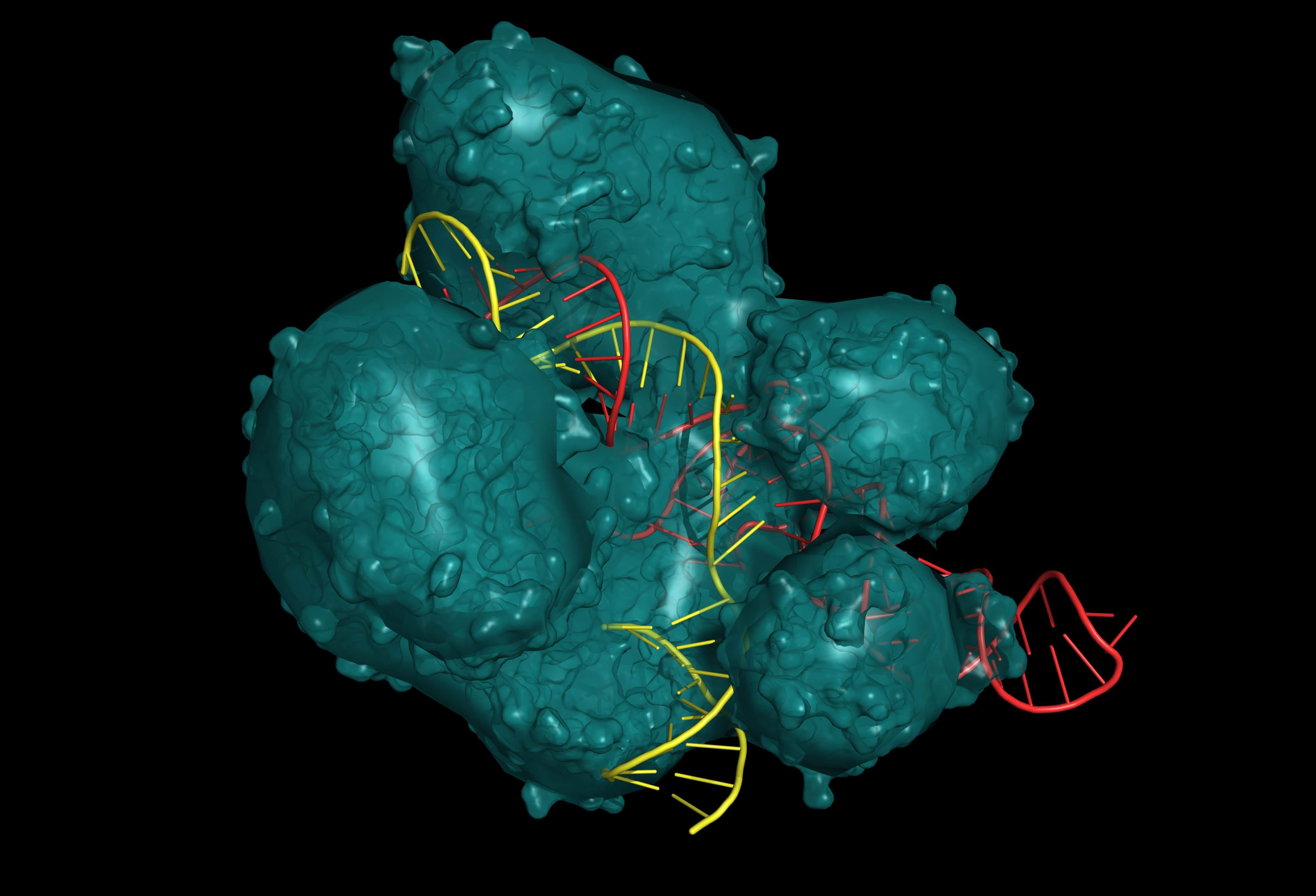
CRISP/Cas9 system for editing, regulating and targeting genomes. The Cas9 protein uses a gRNA (guide RNA) sequence to cut DNA at a complementary site. Protein are in teal, RNA are in red, and DNA are in yellow. (Adobe Stock)
SgRNA sequences in the different cell populations were amplified and deep-sequenced. As each sgRNA is designed to target a specific gene, the genes responsible for changes in SAMHD1 expression can be highlighted. This experiment demonstrated that the IFITM proteins IFITM2 and IFITM3 inhibited SAMHD1 degradation, but IFITM1 did not.
However, the researchers observed that the arms race between viruses continues. “Strikingly, the VSV-G envelope was significantly more sensitive to this IFNα entry block and to IFITMs than HIV-1’s natural envelope,” wrote researchers Ferdinand Roesch, Molly Ohainle and Michael Emerman.
“This highlights important differences between VSV-G pseudotyped and wild-type HIV-1, in particular relative to the pathways they use for viral entry, suggesting that HIV-1 may have evolved to escape restriction factors blocking entry,” note the researchers.
While IFITM proteins blocked the entry of viruses containing the VSV-G capsule protein used to study viral entry in the lab, they were not able to effectively block HIV-1 particles. This means that HIV has further evolved to escape viral restriction factors employed by the human immune system like IFITM proteins.
This work uses Twist Bioscience oligo pools to better understand the arms race between viruses and human cells, specifically in the context of HIV. Twist Bioscience produces high quality oligo pools with a low error rate of 1:1000 nt. By synthesizing the oligo pools on their proprietary silicon platform, Twist Bioscience provides highly uniform sets of oligonucleotides that are evenly represented, allowing the researchers to be sure that important hits are not missed. Additionally, Twist Bioscience’s silicon platform provides oligo pools at an industry leading price in as little as 10 days, meaning researchers can afford to do more and do so faster with their CRISPR experiments.
The results of this research are important for two reasons. Firstly, we now know to be careful when drawing conclusions about HIV penetration into cells when using alternative viral models like VSV-G. Secondly, it allows us to better understand how HIV enters our cells and avoids our immune system, allowing it to propagate. As a result, these findings will aid in the future of HIV research - hopefully one day helping to discover a complete cure for the virus.
Featured image: HIV Viruses attacking cell (Adobe Stock)
What did you think?
Like
Dislike
Love
Surprised
Interesting