Phosphoramidite chemistry is the gold standard method for DNA synthesis that has been used in the industry for almost 35 years. Since its discovery, its simplicity and high efficiency have allowed large volumes of oligonucleotide sequences to be synthesized up to 200 base pairs in length. Currently, it is the only commercially viable chemistry able to provide the volume of DNA required by the synthetic biology market. Here, we take a look at how phosphoramidite chemistry makes the DNA synthesis process so efficient.
On-demand synthetic DNA is essential to the practices of genetics, synthetic biology, and metabolic engineering because it allows researchers to manipulate and better understand life. Creating synthetic DNA requires robust methods that both mimic natural processes and allow for the accurate assembly of custom oligonucleotide sequences.
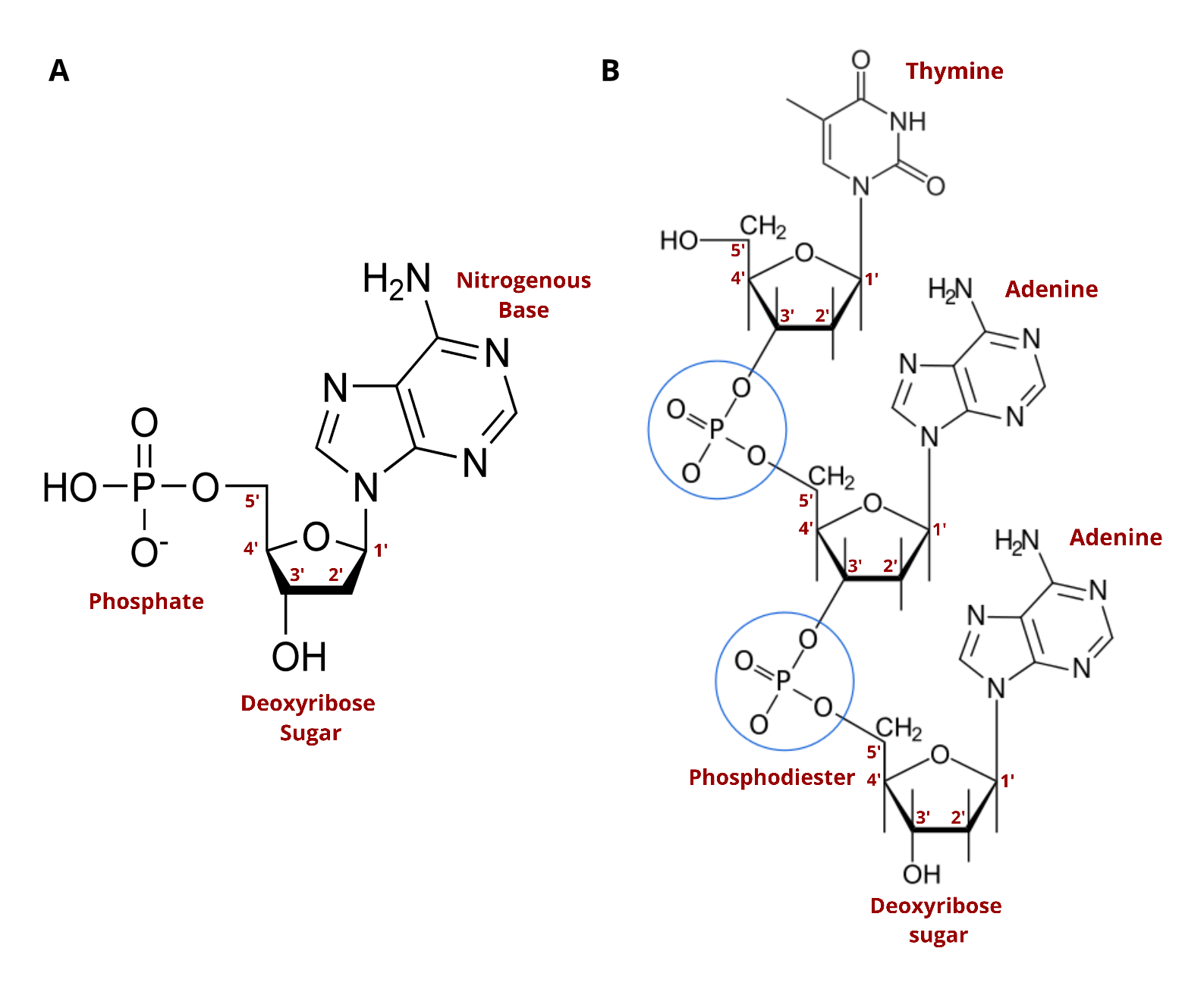
DNA synthesis is therefore one of the most important discoveries of the last century. Natural DNA consists of nucleotides organized into repeating units that form a chemical chain, with each nucleotide linked to another by the action of enzymes. This linkage is enabled by three key chemical components that are found in each nucleotide (see image below).
Methods for creating synthetic DNA mimic this process through the use of a validated technology: phosphoramidite chemistry, which has stood the test of time. In fact, the seminal patent in this space will be celebrating its 37th birthday this year. Here we are going to take a brief look at the standard phosphoramidite chemistry reaction, the properties that make the technology so reliable, and the role it plays in our commercial process at Twist Bioscience.
🧬 Nucleoside vs Nucleotide
You’ll likely see the terms “nucleoside” and “nucleotide” often. The two terms are very similar, with both describing building blocks for DNA and RNA. The only difference is that nucleotides contain a phosphate group in the five prime position which is critical to its ability to form chains. Standard nucleosides can't form chains until they've been modified. Nature phosphorylates nucleosides to create nucleotides which can then string together to form DNA. To make synthetic DNA, nucleosides are turned into nucleoside phosphoramidites.
Phosphoramidite chemistry and oligonucleotide synthesis
Nucleoside Phosphoramidites were first described in 1981. Phosphoramidites are modified nucleosides and are a standard chemical used in modern DNA synthesis. These molecules permit the sequential addition of new bases to the DNA chain in an exquisitely simple and exceptionally efficient cyclic reaction.
Before phosphoramidites, DNA synthesis technologies were limited in the length, quality, and yields of the DNA they produced due to reaction inefficiencies that generated branching, truncated, or mutated oligonucleotide sequences. (For more information on oligonucleotides, see our blog, “How Oligos Changed the World”).
Additionally, growing sequences were predisposed to damage, as the synthesis reaction’s intermediates were highly unstable, and could be broken apart just by exposing them to air. This was largely caused by the inefficient use of extra chemical groups that were added to each nucleotide, called protecting groups. Protecting groups are designed to prevent side reactions that might cause instability in the growing strand.
Phosphoramidites are set apart from these previous methods, in part, by the incorporation of optimal protecting groups.
Protecting groups, DNA, and Phosphoramidite Chemistry
A single natural nucleoside is rich in both hydroxyl groups (-OH) and amino groups (-NH2), both of which can easily interact with many of the other compounds that are used in DNA synthesis. These unintended reactions can result in undesired molecules that do not conform to the characteristic—and functionally important—double-helix structure of DNA.
In 1981, the discovery of the nucleoside phosphoramidite was published, offering a powerful tool for oligo synthesis. As shown below, a standard phosphoramidite molecule contains a phosphite instead of the typical phosphate in nucleotides, as well as four specific protecting groups. These groups provide stable protection, but can be easily removed when necessary. This allows DNA chemists to expose only the reactive centers required to produce a DNA-like molecule, providing a means for tight control over synthesis chemistry. Today, many modified versions of phosphoramidites exist, each with varying properties optimized for specific DNA synthesis processes.
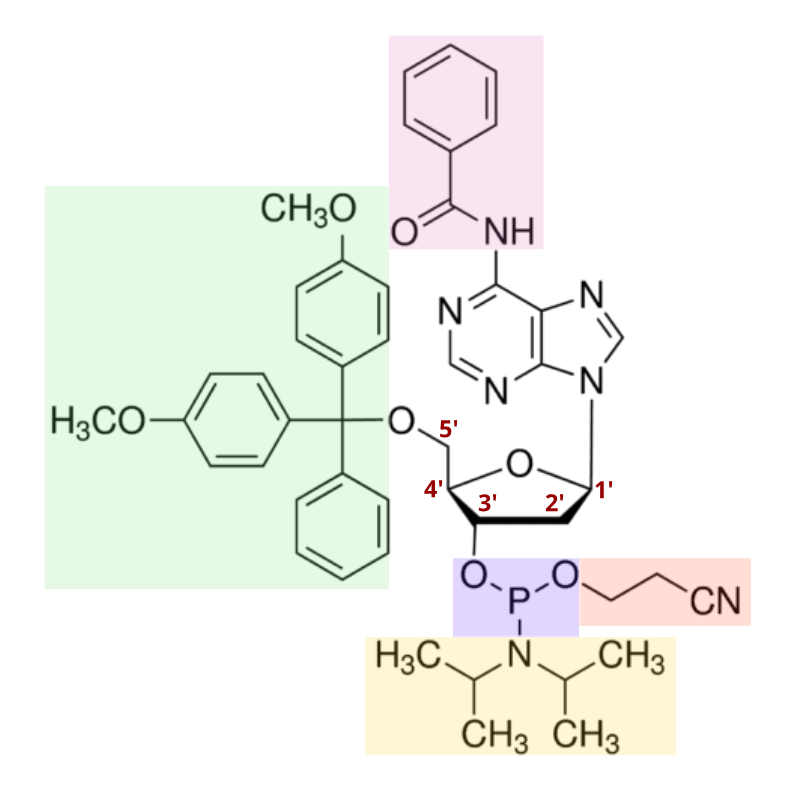
Phosphoramidites and their modified variants remain the gold standard in DNA synthesis technology today. As mentioned in the figure above, the removal of the diisopropylamino group, and the subsequent reaction of the intermediate with the -OH group on a deprotected deoxyribose sugar occurs very quickly. Accordingly, the phosphoramidites then react to link with one another at more than 99% efficiency.
Together, these properties have allowed phosphoramidite chemistry to stand the test of time. And, the first machines that were able to automate oligonucleotide synthesis came to market in the late 1980s, capable of synthesizing a single oligonucleotide in a day. While automation technology has improved and phosphoramidite chemistry is both exceptionally efficient and predictable, manufacturing of DNA has been limited to relatively small-scale operations due to a lack of innovation in the hardware necessary for automation and miniaturization.
Automation and Miniaturization Chemistry
Of note, good chemistry is not enough to make DNA into a tool that serves the throughput required in today’s markets. Synthetic and molecular biology require high volumes of synthetic DNA, often testing thousands of sequence variants at once. Therefore, to serve the needs of this market, we at Twist Bioscience sought to combine phosphoramidite-based chemistry with a proprietary silicon support. We then leverage expertise in miniaturization, engineering, fluid mechanics, robotics, and laboratory information management systems to automate DNA synthesis with extreme precision at an unprecedented scale.
Our use of a silicon support device is a form of solid support DNA synthesis which pre-dates phosphoramidite chemistry. Robert Letsinger—one of the forefathers of modern oligonucleotide synthesis—was the first to synthesize DNA using styrene supports in 1965. Solid-phase chemistry was a major advance in the oligonucleotide synthesis world, as the reaction components could simply be washed away after each new base was added to the oligonucleotide chain. This removed the need for extensive purification of the growing DNA strand at every step.
Silicon makes for an excellent solid support for DNA synthesis. There are many properties of silicon that allow us to miniaturize phosphoramidite chemistry:
- Silicon is flat at microscopic scales, meaning the required fluids in the reaction flow uniformly over the surface.
- Silicon is also a very hard material and other materials can be added or removed without changing the surface.
- Silicon can also be aligned, patterned, and imaged with ease.
- Finally, silicon conducts heat very well, enabling researchers to change the temperature quickly, accurately, and uniformly across the material’s surface.
With our current commercial workflow in place, we are able to synthesize over one million oligonucleotides simultaneously, each with a unique sequence, meaning each can be synthesized for a different product. Typically, oligonucleotides are synthesized at Twist Bioscience on our proprietary silicon platform in a phosphoramidite reaction cycle as seen below:
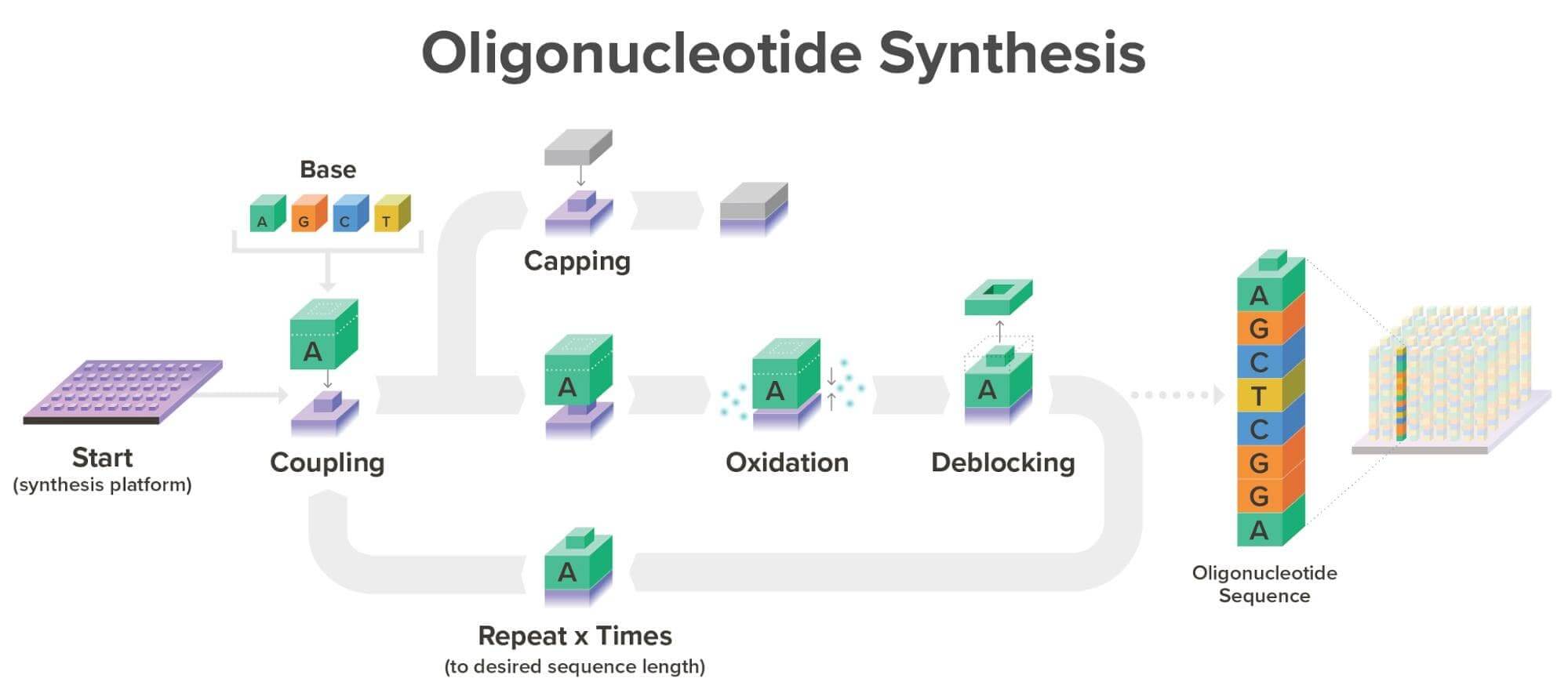
It’s important to note that silicon DNA synthesis technology and phosphoramidite chemistry are not dependent on one another. While phosphoramidite chemistry is the gold standard today, the properties of silicon mean that should another great breakthrough in DNA synthesis technology happen, we can easily adapt our robust commercial infrastructure to incorporate any proven technology that would improve speed, throughput, and/or cost.
We have built a commercial engine to synthesize DNA that enables our customers to change the world.
Already researchers are realizing the benefits of easy access to large-scale DNA synthesis. For example, a team at the University of Ghent recently leveraged Twist’s services to improve their lncRNA sequencing efforts. Elsewhere, in a collaboration with Microsoft Research and the University of Washington, our oligonucleotides are being used as a data storage tool.
We have synthesized new custom double-stranded DNA target capture panels for Next Generation Sequencing, which are being used to better detect deadly viruses. Researchers have utilized our oligonucleotides to generate guide RNA libraries for CRISPR experiments to better understand complex cellular processes.
Additionally, we make gene libraries containing a multitude of variants, from which researchers have been able to identify up to five times the amount of useful molecules compared to other library generation methods—allowing for better protein and antibody engineering
We are committed to driving the cutting edge of new discoveries for health and sustainability through our synthetic DNA. And, we owe much of our success to the beauty of phosphoramidite chemistry.
Cover image: An artist's rendition of the DNA double helix (Adobe Stock)