Discover New Viral Species
The spread of viral outbreaks can have a dramatic impact on our world. The emergence of the SARS-CoV-2 pandemic and epidemics such as the Mpox outbreak highlight the need for improved tools for detecting and surveilling novel viral pathogens. In order to address these, Twist has developed the Comprehensive Viral Research Panel which covers over 3,000 species of known viruses.
This panel's design incorporates the use of the CATCH (Compact Aggregation of Targets for Comprehensive Hybridization) algorithm. For additional details on CATCH, please refer to Metsky HC and Siddle KJ et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nature Biotechnology, 37(2), 160–168 (2019). doi: 10.1038/ s41587-018-0006-x
Learn how one lab is filling an industry need by using Twist's Comprehensive Viral Research Panel.
Discover New Viral Species
The spread of viral outbreaks can have a dramatic impact on our world. The emergence of the SARS-CoV-2 pandemic and epidemics such as the Mpox outbreak highlight the need for improved tools for detecting and surveilling novel viral pathogens. In order to address these, Twist has developed the Comprehensive Viral Research Panel which covers over 3,000 species of known viruses.
This panel's design incorporates the use of the CATCH (Compact Aggregation of Targets for Comprehensive Hybridization) algorithm. For additional details on CATCH, please refer to Metsky HC and Siddle KJ et al. Capturing sequence diversity in metagenomes with comprehensive and scalable probe design. Nature Biotechnology, 37(2), 160–168 (2019). doi: 10.1038/ s41587-018-0006-x
Learn how one lab is filling an industry need by using Twist's Comprehensive Viral Research Panel.
We pulled reference sequences from the RefSeq, FluDB, and VIPR databases to design the 1,052,421 unique probes that make up the Twist Comprehensive Viral Research Panel.
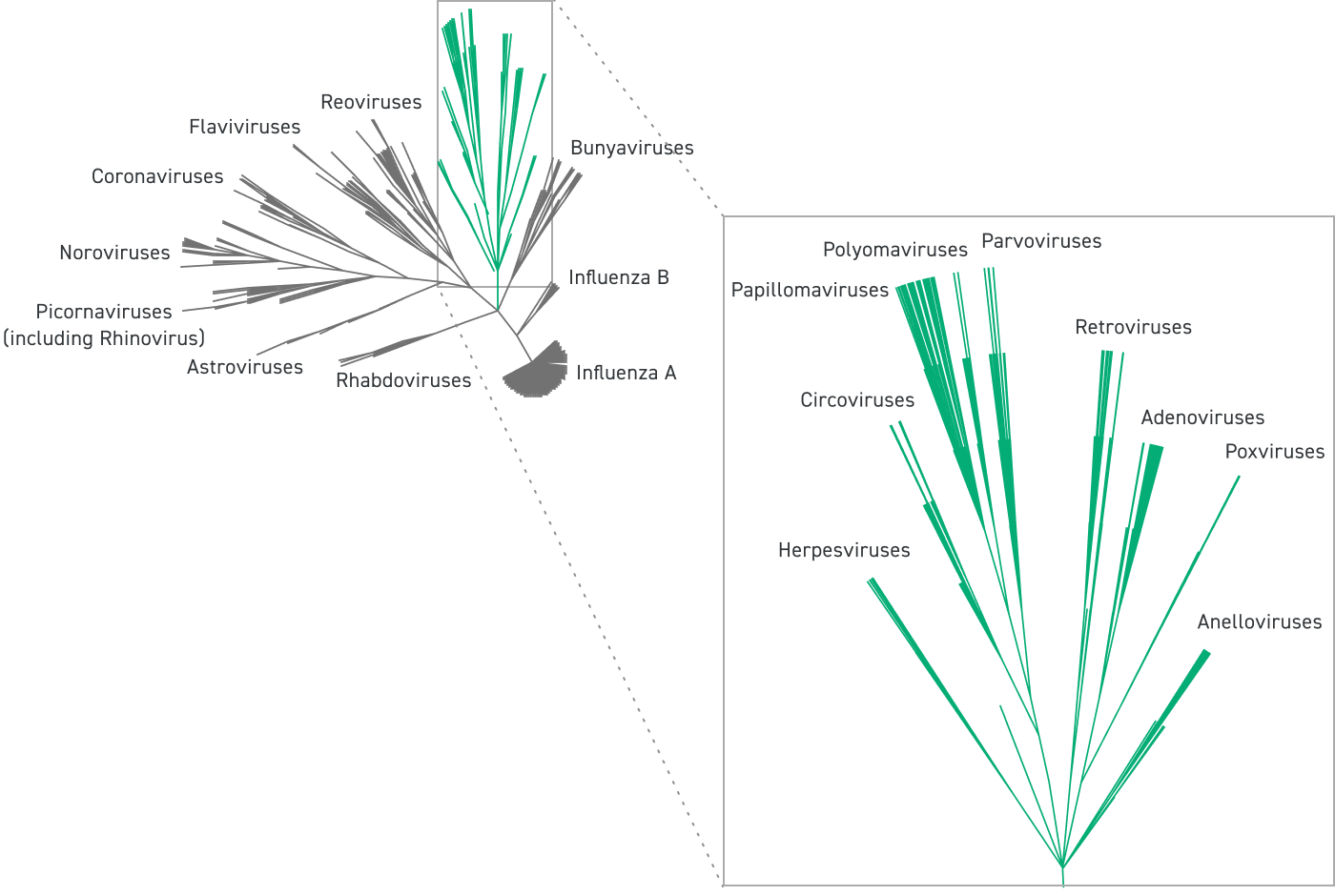
The panel broadly spans every viral family that contains at least one virus known to affect humans, including adenoviruses, coronaviruses, flaviviruses, noroviruses, influenza viruses, and more.
In addition to targeting human-infective viruses, the panel also covers animal viruses (e.g., bat coronaviruses). This enables the discovery of novel viral species regardless of origin.
Its sheer breadth allows the Twist Comprehensive Viral Research Panel to detect novel and evolved viral strains, including those related to a June 2020 outbreak of H1N1 influenza in swine workers.
Natural selection of coronavirus spike and influenza hemagglutinin (HA) glycoproteins in animals facilitated the zoonotic transmission of SARS-CoV-2 and H1N1 influenza to humans.
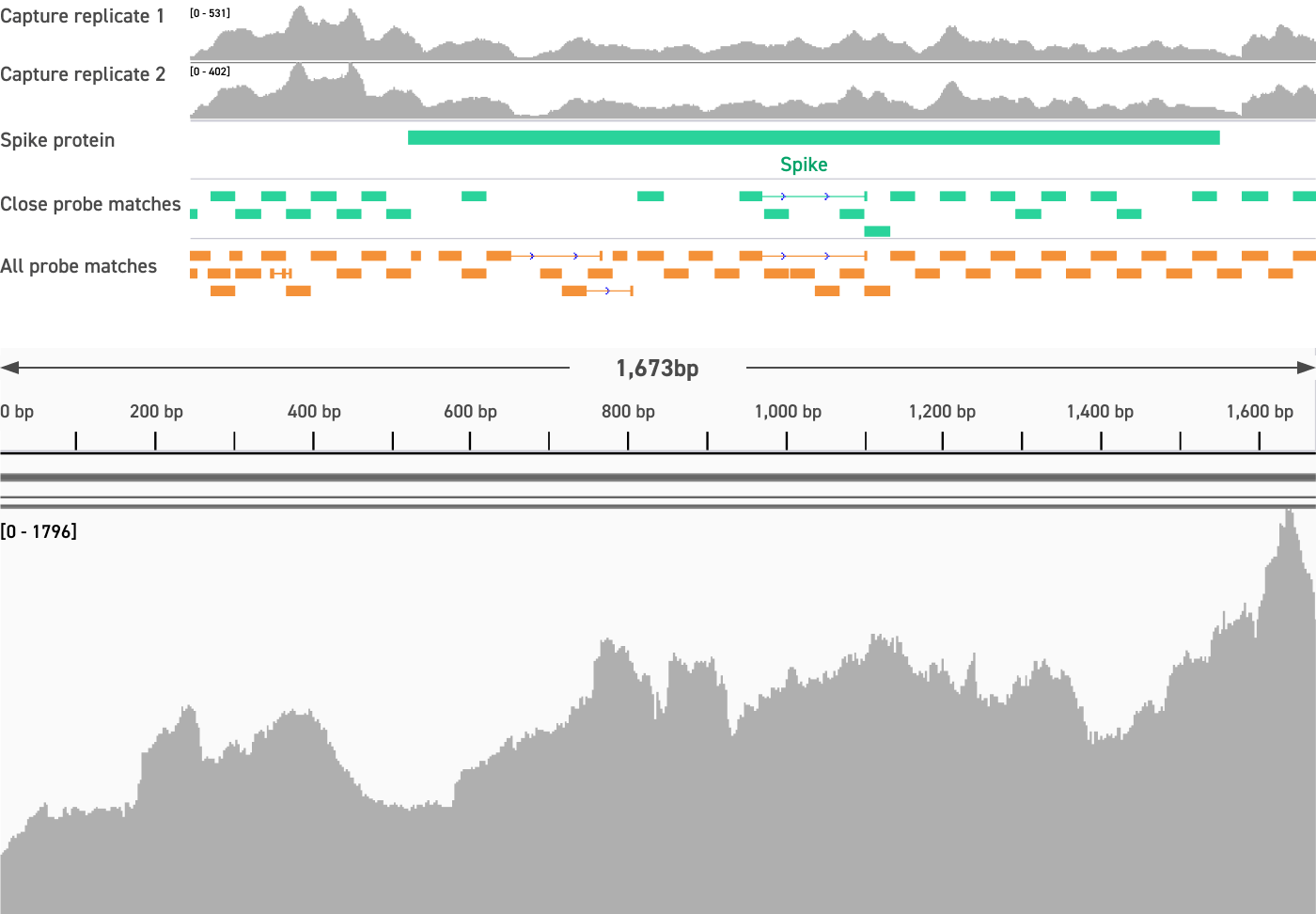
Here, we show that the Twist Comprehensive Viral Research Panel can be used to successfully detect these hard-to-capture sequences, including the spike region of a recently discovered coronavirus (RoBat-CoV GCCDC1 Singapore) and a HA segment isolated from a June 2020 outbreak of H1N1 influenza in swine workers.
Our panel resulted in >99.8% coverage of each sequence to at least 1x depth.
We defined the mismatch sensitivity of the Twist Comprehensive Viral Research Panel by generating a series of HA segments to contain defined levels of sequence variation ranging from 5% to 30% variation from a wild-type HA segment.
Total capture was achieved at 1x coverage in samples with up to 10% sequence variation. Together, these data demonstrate that the Twist Comprehensive Viral Research Panel can capture highly evolved viral sequences.
Multiplex detection of viruses is possible with the Twist Comprehensive Viral Research Panel, enabling metagenomic applications in a variety of sample types.
In a co-infection assay, the panel successfully captured four different virus types (Astrovirus - ssRNA, BK virus - dsDNA, Bocavirus - ssDNA and Picobirnavirus - dsRNA) spiked into human RNA with only 1.2 M reads.
We pulled reference sequences from the RefSeq, FluDB, and VIPR databases to design the 1,052,421 unique probes that make up the Twist Comprehensive Viral Research Panel.
The panel broadly spans every viral family that contains at least one virus known to affect humans, including adenoviruses, coronaviruses, flaviviruses, noroviruses, influenza viruses, and more.
In addition to targeting human-infective viruses, the panel also covers animal viruses (e.g., bat coronaviruses). This enables the discovery of novel viral species regardless of origin.
Its sheer breadth allows the Twist Comprehensive Viral Research Panel to detect novel and evolved viral strains, including those related to a June 2020 outbreak of H1N1 influenza in swine workers.

Natural selection of coronavirus spike and influenza hemagglutinin (HA) glycoproteins in animals facilitated the zoonotic transmission of SARS-CoV-2 and H1N1 influenza to humans.
Here, we show that the Twist Comprehensive Viral Research Panel can be used to successfully detect these hard-to-capture sequences, including the spike region of a recently discovered coronavirus (RoBat-CoV GCCDC1 Singapore) and a HA segment isolated from a June 2020 outbreak of H1N1 influenza in swine workers.
Our panel resulted in >99.8% coverage of each sequence to at least 1x depth.

We defined the mismatch sensitivity of the Twist Comprehensive Viral Research Panel by generating a series of HA segments to contain defined levels of sequence variation ranging from 5% to 30% variation from a wild-type HA segment.
Total capture was achieved at 1x coverage in samples with up to 10% sequence variation. Together, these data demonstrate that the Twist Comprehensive Viral Research Panel can capture highly evolved viral sequences.
Multiplex detection of viruses is possible with the Twist Comprehensive Viral Research Panel, enabling metagenomic applications in a variety of sample types.
In a co-infection assay, the panel successfully captured four different virus types (Astrovirus - ssRNA, BK virus - dsDNA, Bocavirus - ssDNA and Picobirnavirus - dsRNA) spiked into human RNA with only 1.2 M reads.
The Twist Comprehensive Viral Research Panel includes access to the web analysis tool One Codex. In addition to supplying analysis support, this cloud-based tool allows users to document their novel virus discovery efforts with printable reports and publication-ready figures.
One Codex simplifies the analysis of sequence data obtained using the Twist Comprehensive Viral Research Panel. After uploading sequence data with a straightforward drag-and-drop interface, users can generate a report detailing their novel virus discovery results within minutes. Users can also customize the bioinformatics workflow with their own detection thresholds and interpretations.
Purchase of the Twist Comprehensive Viral Research Panel comes with everything needed to start analyzing your results with One Codex, including a coupon code and detailed instructions. Learn more about this streamlined microbial analysis platform at onecodex.com
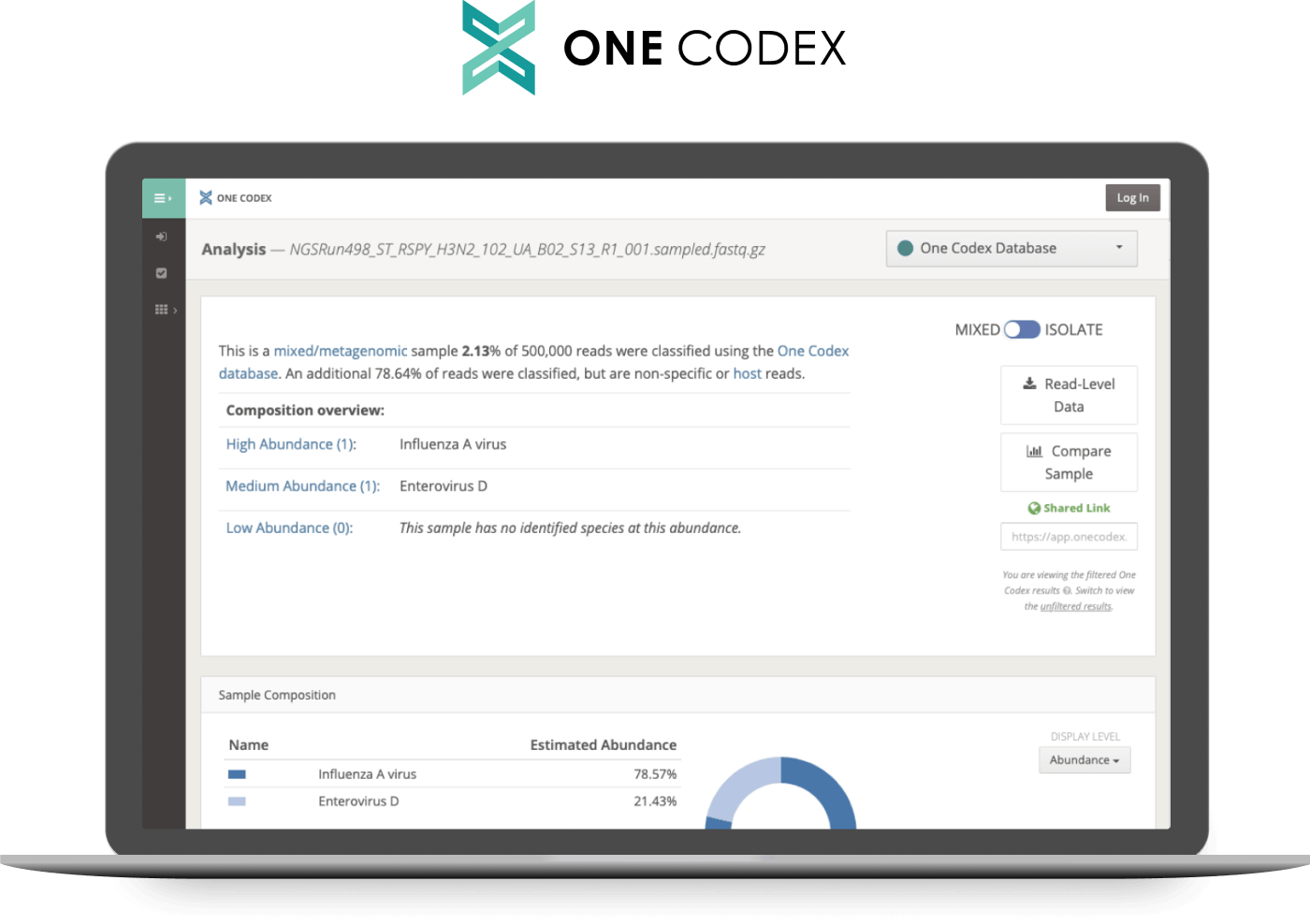
The Twist Comprehensive Viral Research Panel includes access to the web analysis tool One Codex. In addition to supplying analysis support, this cloud-based tool allows users to document their novel virus discovery efforts with printable reports and publication-ready figures.
One Codex simplifies the analysis of sequence data obtained using the Twist Comprehensive Viral Research Panel. After uploading sequence data with a straightforward drag-and-drop interface, users can generate a report detailing their novel virus discovery results within minutes. Users can also customize the bioinformatics workflow with their own detection thresholds and interpretations.
Purchase of the Twist Comprehensive Viral Research Panel comes with everything needed to start analyzing your results with One Codex, including a coupon code and detailed instructions. Learn more about this streamlined microbial analysis platform at onecodex.com

103545
Twist Comprehensive Viral Research Panel with One Codex software, 2 Reactions, Kit103547
Twist Comprehensive Viral Research Panel with One Codex software, 12 Reactions, Kit103548
Twist Comprehensive Viral Research Panel with One Codex software, 96 Reactions, KitWorkflow Step
Target EnrichmentFormat
dsDNAPanel size
36.8 MbContent
Viral sequencesDesign Databases
FluDB, RefSeq, VIPRdbStorage
Store at -5 to -30C103545
Twist Comprehensive Viral Research Panel with One Codex software, 2 Reactions, Kit103547
Twist Comprehensive Viral Research Panel with One Codex software, 12 Reactions, Kit103548
Twist Comprehensive Viral Research Panel with One Codex software, 96 Reactions, KitWorkflow Step
Target EnrichmentFormat
dsDNAPanel size
36.8 MbContent
Viral sequencesDesign Databases
FluDB, RefSeq, VIPRdbStorage
Store at -5 to -30C
Protocol
Twist Total Nucleic Acids Library Preparation Kit for Viral Pathogen Detection and Characterization
Application Note
NGS Target Enrichment of Viral Pathogens using Twist Respiratory Virus Research Panel
Publications
Evaluating metagenomics and targeted approaches for diagnosis and surveillance of viruses
Publications
Targeted whole-viral genome sequencing from formalin-fixed paraffin-embedded neuropathology specimens
Protocol
Twist Total Nucleic Acids Library Preparation Kit for Viral Pathogen Detection and Characterization