Moving Beyond 300 Base Pairs
In a recent episode of the podcast series Lab Talk by The Scientist, guest speaker Josh Tycko’s excitement was infectious. Tycko, a postdoctoral researcher at Harvard Medical School, spoke about how functional genomics is evolving given recent advances in DNA synthesis.
“This is an awesome phase for expressing creativity in science,” he explains, before describing how new DNA synthesis technology is giving researchers access to unprecedented DNA “real estate” for designing screens and uncovering the functions of previously uncharacterized sequences.
Molecular biology advances incrementally, with knowledge gained through years of meticulous labwork. But rapid discovery is possible, typically enabled by underlying technological breakthroughs. For instance, access to next-generation sequencing (NGS) allowed researchers to sequence and annotate genes with remarkable efficiency, speed, and throughput.
When technology simplifies complex processes, creativity can flourish.
We are now in a similar phase with molecular biology as advancements in NGS, artificial intelligence (AI), and DNA synthesis converge. For the first time ever, Twist’s Multiplexed Gene Fragments enable the synthesis of screens up to 500 bp surpassing the previous 300 bp limit. Now, Tycko can explore functional genomics and de novo protein engineering with fewer technical limitations.
“You can just think about your favorite protein family and how you might design a new reporter system,” Tycko describes. “Then using these synthetic libraries a single person, in a single experiment can really map function across an entire protein family in a relatively feasible way.”
What Are Multiplexed Gene Fragments?
Twist Multiplexed Gene Fragments are pools of double-stranded gene fragments, between 301 and 500 bp, that are directly synthesized using massively-paralleled solid phase DNA synthesis. These pools of DNA can contain up to hundreds of thousands of unique DNA sequences, enabling researchers like Tycko to quickly generate a library of candidate DNA sequences or genetic variants for screening.
MGFs have proven transformative, both due to their length and pooled format. Traditional gene fragments are synthesized on a per-fragment basis. This works well for small-scale projects, but becomes costly and complex at scale, limiting massively parallel reporter assays, like those Tycko uses.
As a result, researchers cannot investigate all genetic variants in a single screen, slowing progress and limiting insights. Additionally, standard DNA synthesis maxes out at 300 bp, forcing researchers to stitch multiple DNA fragments together – a complex, laborious process – to create their desired sequence.
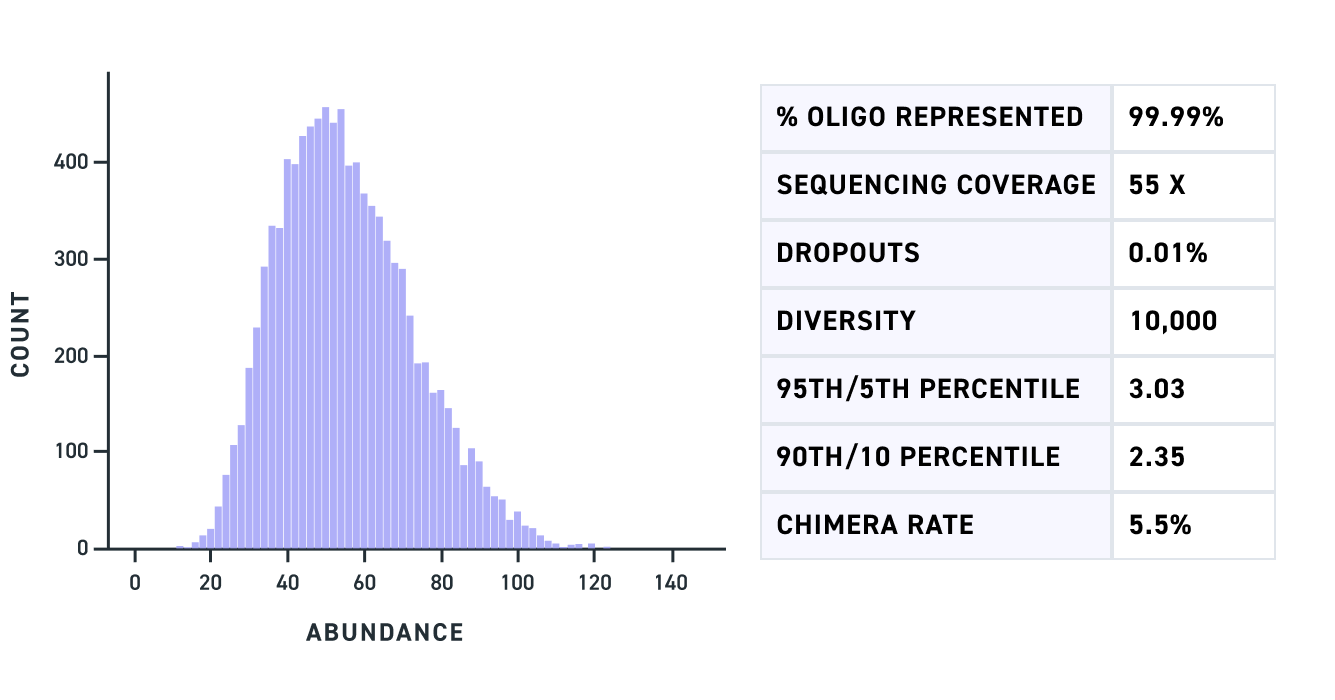
MGFs address these challenges by providing custom pools of double-stranded DNA up to 500 bp at virtually any scale. By synthesizing longer gene fragments in a pooled format, Twist enables Tycko to perform previously unthinkable high-throughput screens.
“Just hearing that MGFs were becoming available is what inspired us,” he said, “we knew that now is the right time to do this high-throughput recruit project.”
High-Throughput Functional Genomics
The low throughput of recruitment assays was a common challenge for Tycko and his colleagues, who study the activity of effector protein domains. “Recruitment assays are an old-school synthetic biology approach,” says Tycko. The method works by engineering a new protein that combines a DNA-binding domain with a transcriptional effector, which, upon target recruitment, influences gene expression.
Though valuable, the traditional assay is low-throughput, requiring synthetic DNA fragments to be pieced together to create novel proteins. Tycko’s team leveraged NGS, automated cell sorting, and MGF to convert this assay into a high-throughput workflow, called HT-Recruit.
HT-Recruit works in the same way as a traditional recruitment assay, only at a larger scale. “We’re testing one candidate effector domain in each cell,” he explained. “But, we’re growing millions, or hundreds of millions of cells at a time.” This allows Tycko’s team to greatly increase the number of effector domains being tested per experiment.
“The most enabling research tool for HT-Recruit was the new development of oligo libraries that contain longer pieces of DNA,” he explained. Longer gene fragments encode for longer protein domains. Since 30% of known gene effector domains are 100 amino acids long, they’ve been excluded from studies because they rely on gene fragments over 300 bp.
In vivo, protein domains interact in complex ways that influence gene expression, but, due to gene fragment size limitations, studying combinatorial effector domains has been a challenge.
“Before MGFs,” Tycko clarifies, “I needed to come up with ways to clone libraries where we made one domain at a time, fused them together with some type of linker, and studied their function using those types of combinatorial libraries…It’s definitely a more difficult undertaking.”
Now with access to MGF, Tycko can use HT-Recruit for massively parallel combinatorial testing of effector domains. “The ability to just type into the computer the combination of domains that are interesting and just have them printed out in one go, it makes that process a lot easier.”
Discovering The Unexpected
With greater sequence space and the potential for high-throughput testing, MGF enables researchers to screen and test new hypotheses with greater speed and freedom. “The number of questions you can ask using synthetic libraries is probably boundless,” Tycko emphasized. “It opens you up to not only pursuing a hypothesis about your favorite protein family, but also opens you up to discovering the unexpected.”
”It opens you up to discovering the unexpected”
In 2020, he and his colleagues leveraged HT-Recruit to test all known KRAB protein domains, uncovering that while most act as repressors, some unexpectedly function as strong activators1.
Such an unexpected discovery not only helps to shed light on the evolution of effector domains, but has also led to the development of a novel, more potent CRISPRa complex2. “That was just a really cool path that I think we wouldn’t have gone down without having the MGF library.”
One Example Among Many
Tycko’s work highlights how MGFs can be a transformative tool in functional genomics. Rather than leaving a potential variant on the cutting floor, unexamined, researchers now can test expansive variant libraries rapidly and efficiently.
This frees you up to be creative, find answers quickly, and discover the unexpected.
References
- Tycko, Josh, et al. “High-Throughput Discovery and Characterization of Human Transcriptional Effectors.” Cell, vol. 183, no. 7, Dec. 2020, pp. 2020-2035.e16, https://doi.org/10.1016/j.cell.2020.11.024.
- Tycko, Josh, et al. “Development of Compact Transcriptional Effectors Using High-Throughput Measurements in Diverse Contexts.” Nature Biotechnology, 1 Nov. 2024, https://doi.org/10.1038/s41587-024-02442-6.
What did you think?
Like
Dislike
Love
Surprised
Interesting