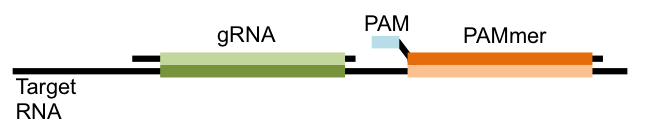December 12, 2017
5 min read
New Research Showcase: Editing RNA with CRISPR
Researchers use guide RNA (gRNA) to chaperone the DNA cutting enzyme Cas9 to specific regions in the genome. Cas9 then makes a double-stranded cut in the DNA, which can become...
There is no question that CRISPR is the hottest technology in genome editing right now. Its power comes from its simplicity. Researchers use guide RNA (gRNA) to chaperone the DNA cutting enzyme Cas9 to specific regions in the genome. Cas9 then makes a double-stranded cut in the DNA, which can become the site for insertion of new genetic material – or with a second cut, facilitates gene deletion.
Cas9 has exploded onto the scene, with exponential growth in research using this system since its discovery in 2012. Some high impact studies nestled amongst these articles include proof-of-principle disease cures, new models for studying genetic disorders, faster animal model generation, theories for the rapid eradication of malaria from mosquitos, controversial human germ line modification, and systems to highlight new drug targets for cancer.
While its simplicity has changed the face of engineering biology, sometimes the CRISPR system makes unwanted mistakes. So called off-target effects occur because the 20-nucleic-acid-long gRNA could chaperone Cas9 to more than one position in the genome. This, combined with Cas9’s ‘relaxed’ nature – not all of the nucleic acids in the gRNA always need to be complimentary with the DNA sequence of interest to produce a cut – can lead to occasional unintended errors.
On March 17th 2016, Gene Yeo’s research group at the University of California offered a colossal breakthrough. Instead of recognizing double-stranded DNA, his team’s RCas9 specifically recognizes single-stranded RNA transcribed from the genome. You can read the paper here.
Why Targeting RNA is Important
The design of Yeo’s RNA targeting system constrains Cas9 into binding an RNA sequence only, not DNA. This occurs by virtue of two guide RNA components which have to simultaneously come together – each with recognition for the RNA sequence – in order for RCas9 to associate with the sequence (elaborated in ‘the science’ box if you’re interested).
RCas9’s specificity for only the transcriptome could unlock protein editing at the RNA level, and not the genome level. This is important as only around 85% of the genome is transcribed (with only 2% being protein coding), meaning RCas9 has less genetic sequence that it could aberrantly bind compared to the conventional Cas9 system. Additionally it is worth mentioning that each RNA is ‘transient’ – it is rapidly recycled by the cell, whereas a DNA sequence remains once changed. As off-target effects are considerably rare compared to on-target effects, it is highly likely that any off-target effect will be quickly diluted out of the cell environment as the erroneous sequences are recycled by the cell’s natural machinery.
Temporary mutations can also be introduced into proteins by RCas9 without touching the genome. One could imagine RCas9 treatments or cures for currently untreatable diseases that do not require modification of the genome, by destroying viral RNA, transcripts from bacteria, or even cancer-only gene transcripts. An example application could be degrading the product of the Bcr-Abl tyrosine kinase fusion protein that causes around 90% of chronic leukemia. In this case, a tyrosine kinase protein (which is usually turned off in cells) is turned on permanently – causing the disease. Targeting this transcript could suppress the disease, and would be as simple to adapt as changing gRNA sequence if treatment resistance were to arise. By contrast, conventional pharmaceutical compounds would take years of molecule discovery.
Also, a diseased cell line expressing RCas9 could easily have numerous proteins perturbed simultaneously, simply by injecting numerous guide RNA constructs into the cell. This will be important for studying diseased cells, as only one cell line needs to be created to model hundreds of phenotypes.
RCas9’s application does not end here. Yeo’s study showed that a non-cutting RCas9 fused to a protein that emits red light could be used to track the movement of RNA around the cell. This is a huge improvement on previous technologies used to track RNA movement, and will be important in the study and treatment of RNA-based diseases like myotonic dystrophy.
RCas9 is a tremendous breakthrough for the broadened, safe application of CRISPR technologies, and we are excited to see where it could take genome science in the next few years!
Science Deep-Dive
Previous studies showed that in the test tube Cas9 could target RNA sequences if another guiding RNA sequence, called a PAMmer, was also used alongside the gRNA. This PAMmer is complimentary to the RNA sequence that is next to the sequence recognized by the gRNA. It also contains a component called a protospacer adjacent motif (PAM) that is essential for Cas9 to make its cut (see Figure).
In conventional CRISPR experiments, Cas9 is targeted to a genome sequence that has this PAM sequence next to it. Instead, a totally PAM-free RNA sequence is targeted. Yeo’s team moved this system from the test tube to living cells, showing that a non-cutting Cas9 fused to a fluorescent protein can localize in the nucleus of human cells when a gRNA complimentary for a human-encoded RNAs is added. When a gRNA for an untranscribed human RNA was added, Cas9 did not localize to the nucleus.
To show this was no fluke, Yeo’s group also showed that the same system could be used to track the movement of RNA around the cell in real time. RNA transcripts known to localize to granules that form when a cell is under oxidative stress could be tracked as they moved from the nucleus to the granules.
Yeo’s group also tested the effect on translated protein levels and the localization of the transcripts – two important factors for the normal, healthy functioning of a cell. It was shown that when subject to Cas9 RNA binding, there was no difference in either factor compared to non-targeted cells – indicating that it is minimally invasive on cell function, and a perfect platform on which to build for the study of cellular disease.
What did you think?
Like
Dislike
Love
Surprised
Interesting
Read Next
Get the latest by subscribing to our blog