Explore VHH sequence space with three new single domain antibody libraries.
VHH Ratio
Specific oligo pools model the natural VHH repertoire
VHH Shuffle
Natural llama CDR sequences in the context of a llama consensus framework
VHH hShuffle
Natural llama CDR sequences in the context of a partially humanized VHH framework
VHH hShuffle HI
Unique method shuffles millions of llama and human CDR sequences within the context of a partially humanized VHH framework.
VHH hShuffle GPCR
Close to over 100K GPCR binding motifs are taken from the GPCR 2.0 library and incorporated in the VHH hShuffle CDR3.
hCamel Bactrian
Disulfide in H1-H3 for enhanced stability. Over 25 Unique VHH Clones discovered against CD70 from two 384w plates.
hCamel Zero
Engineered to have zero Cys in CDRs and additional diversity in CDR1 and CDR2. Over 67 Unique VHH Clonesd discovered against Omicron S1 from two 384w plates.
Explore VHH sequence space with three new single domain antibody libraries.
VHH Ratio
Specific oligo pools model the natural VHH repertoire
VHH Shuffle
Natural llama CDR sequences in the context of a llama consensus framework
VHH hShuffle
Natural llama CDR sequences in the context of a partially humanized VHH framework
VHH hShuffle HI
Unique method shuffles millions of llama and human CDR sequences within the context of a partially humanized VHH framework.
VHH hShuffle GPCR
Close to over 100K GPCR binding motifs are taken from the GPCR 2.0 library and incorporated in the VHH hShuffle CDR3.
hCamel Bactrian
Disulfide in H1-H3 for enhanced stability. Over 25 Unique VHH Clones discovered against CD70 from two 384w plates.
hCamel Zero
Engineered to have zero Cys in CDRs and additional diversity in CDR1 and CDR2. Over 67 Unique VHH Clonesd discovered against Omicron S1 from two 384w plates.
- 2391 CDR sequences analyzed for position-specific variation
- Controlled CDR diversity introduced in the library
- Consensus llama framework
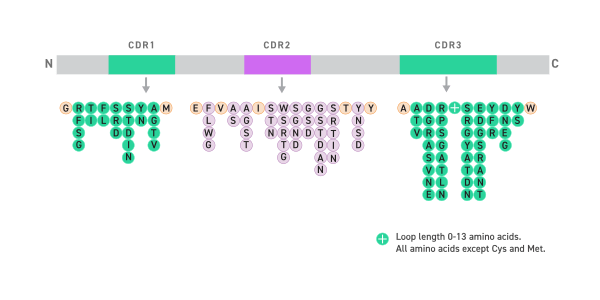
- Each unique CDR individually synthesized
- Shuffled in consensus llama framework
- Final diversity of library > theoretical diversity
- Theoretical library diversity of 3.2 × 10^9
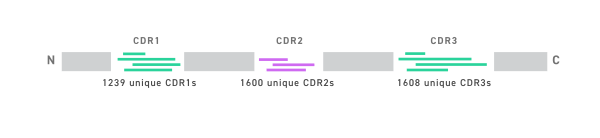
- Shuffled CDRs with theoretical library diversity of 3.2 × 109
- Partially humanized framework: Framework 1, 3 and 4 were
humanized using the human germline DP-47 framework
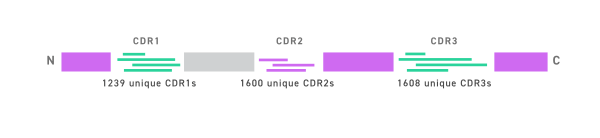
- 2391 CDR sequences analyzed for position-specific variation
- Controlled CDR diversity introduced in the library
- Consensus llama framework

- Each unique CDR individually synthesized
- Shuffled in consensus llama framework
- Final diversity of library > theoretical diversity
- Theoretical library diversity of 3.2 × 10^9

- Shuffled CDRs with theoretical library diversity of 3.2 × 109
- Partially humanized framework: Framework 1, 3 and 4 were
humanized using the human germline DP-47 framework
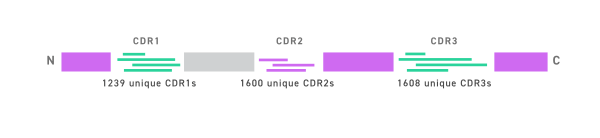
1 × 384 well plate picked per library per round
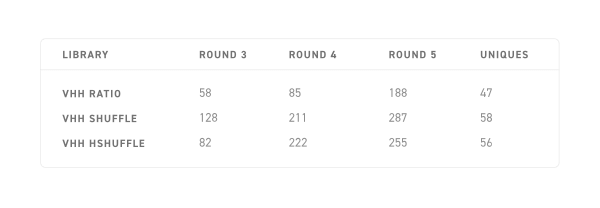
Out of 140 VHH binders
- 51 variants < 100 nM
- 90 variants < 200 nM
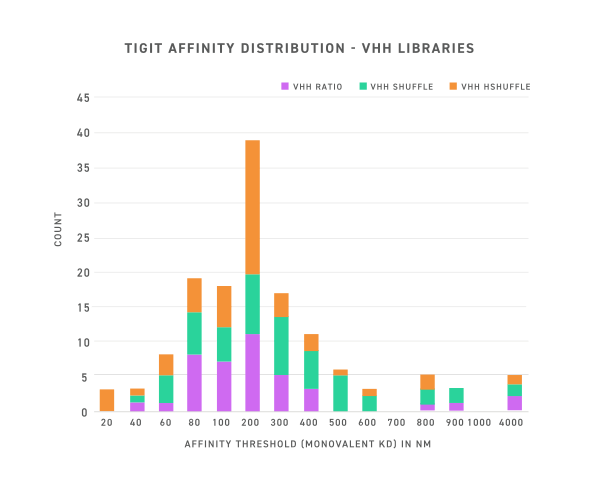
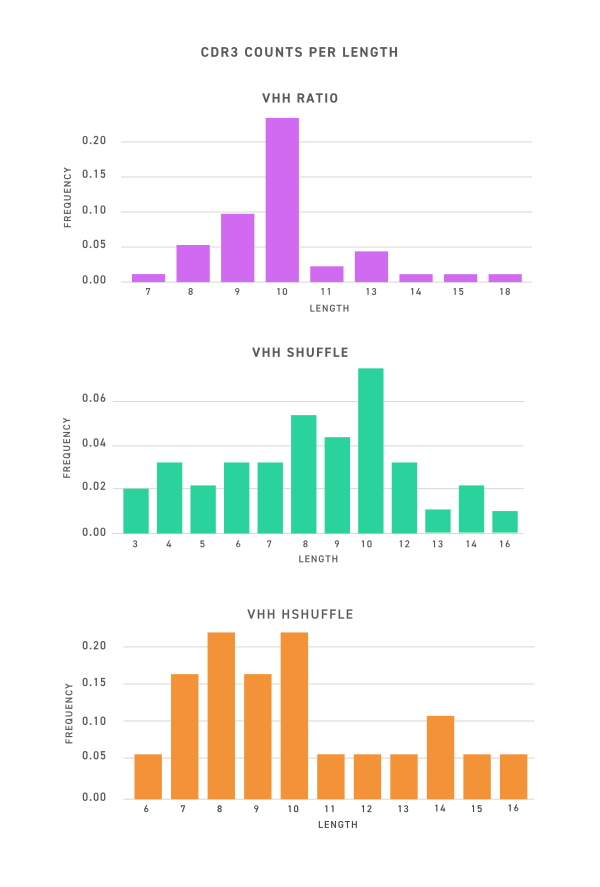
Anti-TIGIT clones from VHH libraries encompass a range of affinities and diversity
1 × 384 well plate picked per library per round

Out of 140 VHH binders
- 51 variants < 100 nM
- 90 variants < 200 nM

Anti-TIGIT clones from VHH libraries encompass a range of affinities and diversity
