Twist Bioscience HQ
681 Gateway Blvd
South San Francisco, CA 94080
Preparing Your Order
- What are the requirements for my custom vector?
- On the Custom Vector On-boarding app, do I need to provide the complete vector only sequence?
- Do I need to add restriction sites to the ends of my Insert Sequence?
- How much DNA is required when submitting a custom vector?
- On the Custom Vector On-boarding app, can I submit just the sequence of the multiple cloning site?
What are the requirements for my custom vector?
Ensure your vector meets the following requirements:
- Vector Volume ≥ 100 uL (Plasmid material cannot be submitted on filter paper)
- Vector Concentration ≥ 100 ng/uL
- Vector Mass 10 ug
- Must have bacterial origin of replication (not low copy)
- Must be clonal population
- Must be circular
- Must have antibiotic resistance marker
- Must not contain homopolymers/repetitive sequence >9bp within 40bp of the insertion site
- Repeats: Must contain at least 40bp of unique sequence with no direct or inverted repeats >19bp upstream and downstream of the insertion site
- Must not contain ccdB in final gene product
- Must be able to linearize by Restriction Enzyme site within 50bp of insertion site
- Must be BSL-1 compliant
Antibiotic Resistance:
Plasmid must contain one of the below antibiotic resistance markers for E.coli. Standard concentrations are below:
|
Antibiotic |
Standard concentration available in Production (ug/mL) |
|
Kan |
50 |
|
AMP |
100 |
|
CHLOR |
25 |
|
APRA |
50 |
|
SPEC |
100 |
|
TET |
10 |
|
ZEO |
50 |
|
BLAST |
100 |
Cell Lines (E. coli)
- Cell Line available in Production
- Cloning strain similar to DH10Beta
- Stbl3-like strain (can be grown at 37℃)
Copy number
Only constitutive origins of replication are allowed.
|
Twist Definition |
Accepted |
Example |
|
High |
Yes |
ColE1/pMB1/pBR322/pUC |
|
Medium |
Yes, reduced miniprep yield |
p15A |
|
Low |
No |
pSC101 |
Growth Conditions
- Standard Temperature: 37℃/30℃
- Standard Time: Overnight/24hrs
- Plated colonies must reach an automation friendly size after 16hrs/24hrs of growth on LB-Lennox.
Still have questions? Contact Us
On the Custom Vector On-boarding app, do I need to provide the complete vector only sequence?
You will be required to upload the complete vector sequence in GenBank format with the full sequence of the plasmid that will be shipped to Twist. Twist will sequence the incoming vector to ensure the sequences match. Any discrepancies will delay the on-boarding process.
Still have questions? Contact Us
Do I need to add restriction sites to the ends of my Insert Sequence?
We do not use restriction enzyme based cloning. Please do not include any extra nucleotides in your inserts in anticipation of Restriction Enzyme digestion followed by DNA ligation.
When you place an order for a clonal gene, we synthesize exactly the insert sequence you send us. If you do include flanking Restriction Enzyme sites, we will synthesize those, too; extra base pairs are not excised. The insert sequences are not processed in any way once they are received.
If you are placing a clonal gene order using a Twist or Custom Vector, we highly recommend that you double check the sequence of the final construct using the Download sequences button in the designer:
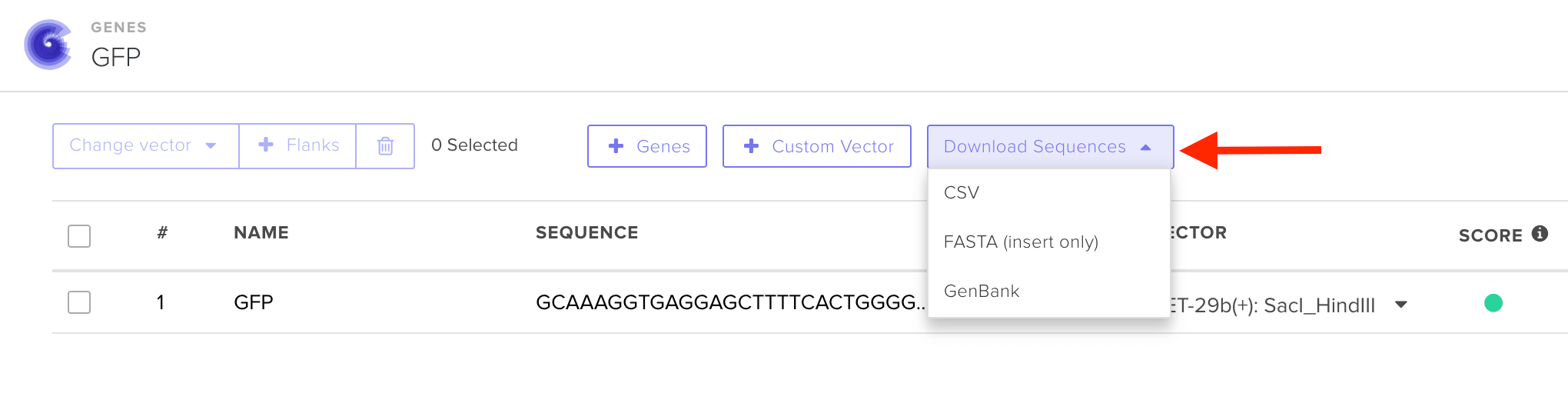 Please note that once a gene order has been placed, we cannot alter the sequence in any way - if you discover an error in your construct, please contact customersupport@twistbioscience.com for assistance.
Please note that once a gene order has been placed, we cannot alter the sequence in any way - if you discover an error in your construct, please contact customersupport@twistbioscience.com for assistance.
Still have questions? Contact Us
How much DNA is required when submitting a custom vector?
Please provide us with 10 ug vector DNA at a concentration of 100 ng/ul in at least 100 ul. Insufficient quantity of vector DNA will cause a delay to the onboarding process.
Still have questions? Contact Us
On the Custom Vector On-boarding app, can I submit just the sequence of the multiple cloning site?
No, you must include the complete sequence of the vector sent to Twist. We use this information for sequence QC and to confirm the cloning strategy. We will sequence your vector and compare it to the sequence you sent us. Errors or mismatches between the two sequence results may delay vector onboarding.
Still have questions? Contact Us
What are the requirements for my custom vector?
Ensure your vector meets the following requirements:
- Vector Volume ≥ 100 uL (Plasmid material cannot be submitted on filter paper)
- Vector Concentration ≥ 100 ng/uL
- Vector Mass 10 ug
- Must have bacterial origin of replication (not low copy)
- Must be clonal population
- Must be circular
- Must have antibiotic resistance marker
- Must not contain homopolymers/repetitive sequence >9bp within 40bp of the insertion site
- Repeats: Must contain at least 40bp of unique sequence with no direct or inverted repeats >19bp upstream and downstream of the insertion site
- Must not contain ccdB in final gene product
- Must be able to linearize by Restriction Enzyme site within 50bp of insertion site
- Must be BSL-1 compliant
Antibiotic Resistance:
Plasmid must contain one of the below antibiotic resistance markers for E.coli. Standard concentrations are below:
|
Antibiotic |
Standard concentration available in Production (ug/mL) |
|
Kan |
50 |
|
AMP |
100 |
|
CHLOR |
25 |
|
APRA |
50 |
|
SPEC |
100 |
|
TET |
10 |
|
ZEO |
50 |
|
BLAST |
100 |
Cell Lines (E. coli)
- Cell Line available in Production
- Cloning strain similar to DH10Beta
- Stbl3-like strain (can be grown at 37℃)
Copy number
Only constitutive origins of replication are allowed.
|
Twist Definition |
Accepted |
Example |
|
High |
Yes |
ColE1/pMB1/pBR322/pUC |
|
Medium |
Yes, reduced miniprep yield |
p15A |
|
Low |
No |
pSC101 |
Growth Conditions
- Standard Temperature: 37℃/30℃
- Standard Time: Overnight/24hrs
- Plated colonies must reach an automation friendly size after 16hrs/24hrs of growth on LB-Lennox.
Still have questions? Contact Us
On the Custom Vector On-boarding app, do I need to provide the complete vector only sequence?
You will be required to upload the complete vector sequence in GenBank format with the full sequence of the plasmid that will be shipped to Twist. Twist will sequence the incoming vector to ensure the sequences match. Any discrepancies will delay the on-boarding process.
Still have questions? Contact Us
Do I need to add restriction sites to the ends of my Insert Sequence?
We do not use restriction enzyme based cloning. Please do not include any extra nucleotides in your inserts in anticipation of Restriction Enzyme digestion followed by DNA ligation.
When you place an order for a clonal gene, we synthesize exactly the insert sequence you send us. If you do include flanking Restriction Enzyme sites, we will synthesize those, too; extra base pairs are not excised. The insert sequences are not processed in any way once they are received.
If you are placing a clonal gene order using a Twist or Custom Vector, we highly recommend that you double check the sequence of the final construct using the Download sequences button in the designer:
 Please note that once a gene order has been placed, we cannot alter the sequence in any way - if you discover an error in your construct, please contact customersupport@twistbioscience.com for assistance.
Please note that once a gene order has been placed, we cannot alter the sequence in any way - if you discover an error in your construct, please contact customersupport@twistbioscience.com for assistance.
Still have questions? Contact Us
How much DNA is required when submitting a custom vector?
Please provide us with 10 ug vector DNA at a concentration of 100 ng/ul in at least 100 ul. Insufficient quantity of vector DNA will cause a delay to the onboarding process.
Still have questions? Contact Us
On the Custom Vector On-boarding app, can I submit just the sequence of the multiple cloning site?
No, you must include the complete sequence of the vector sent to Twist. We use this information for sequence QC and to confirm the cloning strategy. We will sequence your vector and compare it to the sequence you sent us. Errors or mismatches between the two sequence results may delay vector onboarding.
Still have questions? Contact Us







